Amylases
There are three types of amylases: alpha, beta, and gamma. All can be present in bacteria, although they each have preferred environments. Alpha amylase is a major digestive enzyme in animals and works optimally at a pH of 6.7-7.0. Beta amylase is present in fruit and microbes. Gamma amylase is best in environments with a pH of around 3.0.
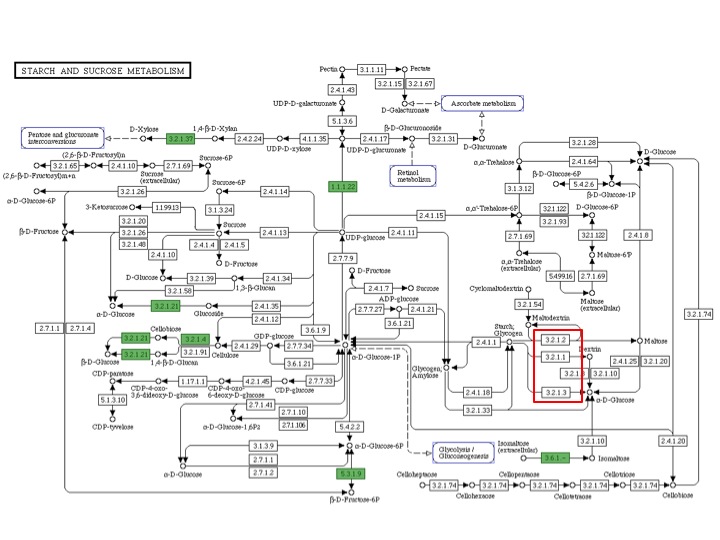
In H. utahensis, I found both alpha and gamma amylases. Alpha amylase is from bps 1748453-1750477. A BLAST of this DNA segment with proteins in NCBI's database matches it to an alpha amylase from Haloquadratum walsbyi with an e-value of 7x10^-172. Gamma amylase is from bps 1751071-1755111. A BLAST of this DNA segment with NCBI's protein database matches it to a glucoamylase (another name for gamma amylase) from Haloarcula marismortui with an e-value of 0.0. Despite these findings, the RAST KEGG Pathway annotation did not identify any amylase genes. Here is an image of the KEGG pathway annotation. Enzymes identified are in boxes with a green background. Amylase genes are boxed in red. Alpha amylase is 3.2.1.1 and gamma amylase is 3.2.1.3.
KEGG pathway annotation also did not indicate RAST found any enzymes whose substrates or products were dextrin or alpha-D-glucose. However, I found a possible 4-alpha-glucanotransferase (EC 2.1.4.25) when BLASTing the sequence for this enzyme from Burkholderia vietnamiensis G4 with our genome with an e value of 8e-21. I also found this enzyme listed on the RAST spreadsheet from bps 1746766 to 1745279. I also found a potential oligo-1,6-glucosidase (EC 3.2.1.10) when BLASTing the sequence for this enzyme from Lactobacillus salivarius UCC118 with our genome with an e value of 7e-24. However, there were no oligo-1,6-glucosidases reported on the RAST spreadsheet.