Determining whether genes called in JGI and RAST are identical
When working with multiple annotations, it is often useful to determine if a gene called by one annotation service is called by another annotation service. The easiest way to determine if genes are called in both annotations is to compare start and stop codons. For large numbers of genes, Perl programs can be created to automate the process of comparing annotated genomes. However, if only a small number of genes are being compared, it can often be more efficient to compare start and stop codons manually.
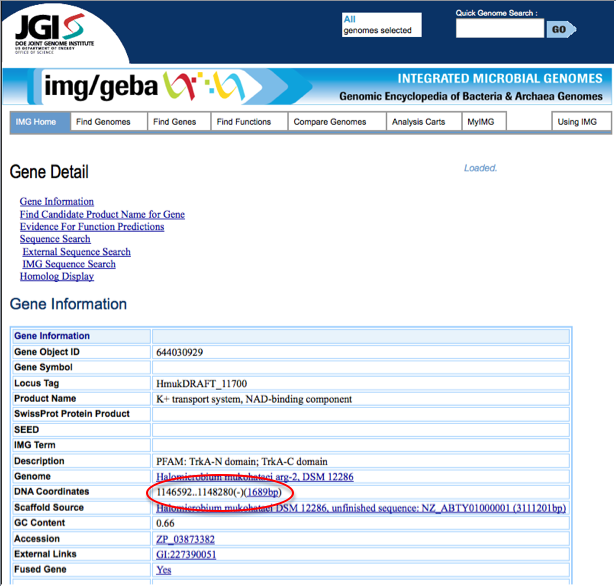
To compare start and stop codons from JGI and RAST, first one must find the start and stop codons for the gene of interest. In JGI, start from the Gene detail page. On this page, under Gene Information, the start and stop codon are listed next to DNA coordinate. JGI also labels whether the gene is on the plus strand or minus strand of DNA. The start and stop codons for a sample gene from JGI are circled in red below.
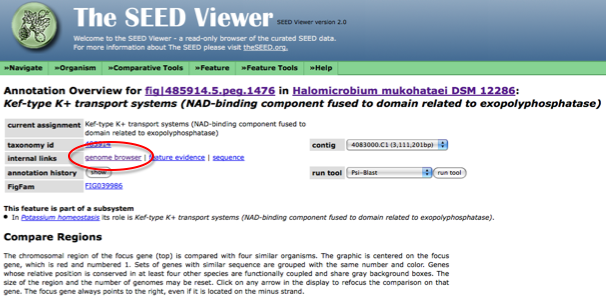
To find the start and stop codon for a gene in RAST, first browse the genome using the SEED viewer. Once you have selected your gene of interest, go to it's [Annotation overview page]. Once on this page, next to internal links select the genome browser (see figure below).
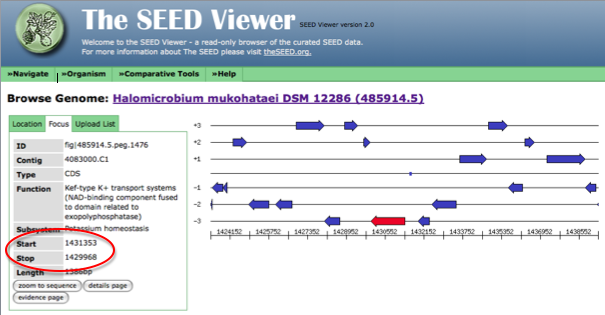
After clicking on the genome browser link, the stop and start codon for your gene will be displayed as shown in the figure below.
Notice that RAST does not specify whether the gene is on the + or - DNA strand. Instead, the order of the start and stop codons listed varies depending on which strand the gene is found on. Because of this fact, it is necessary to compare start and stop codons from RAST to both the start and stop codon called in JGI to determine if the genes are identical or similar. The start and stop codon may be backwards in JGI compared to RAST. Additionally, if either the start or stop codon varies between these annotations, failure to compare both start and stop codons could cause you to miss finding a gene that similar between the two annotation sites but not identical.
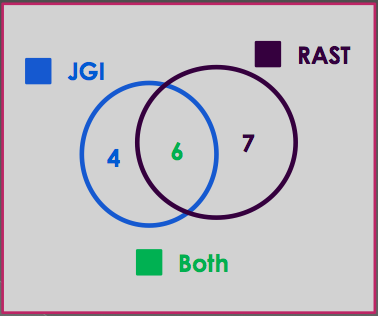
This method of gene comparison is most helpful when one is investigating a finite list of genes of similar function. For example, I did a project that involved genes related to potassium homeostasis. After doing a search in both JGI and RAST, I found 10 genes related to potassium in JGI and 13 genes in RAST. I used the method described above to determine which genes were similar or identical in both annotations. I found 6 genes that were called in both JGI and RAST. Four genes were only called in JGI and seven genes were only called in RAST. See the figure below for clarification of these results.
Perhaps if our genome's Manatee annotation comes back soon, I can also add a section about finding the start and stop codon for this annotation service.
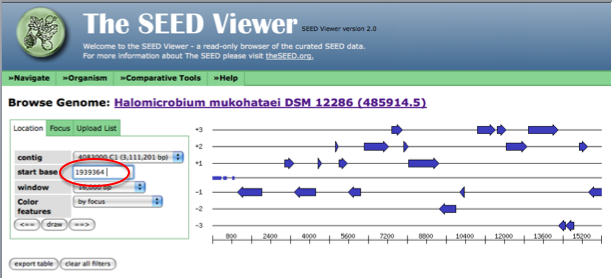
There are other ways to compare called genes from different annotations that do not lend themselves to the above method. For example, if you find a gene of interest in JGI and would like to determine if this gene exists in RAST, you would need to know the gene name in the RAST system to use the start/stop codon method. Because different annotation sites utilize different nomenclature systems, it is unlikely that one would know the name of the gene of interest in the RAST system when the only information available is that from JGI. In a case such as this, RAST has a genome browser for the whole organism that is helpful. In the toolbar on the top of the SEED view page, hover over organism until a drop down menu appears. From the menu, select genome browser. For the start base section, enter the start codon value for your gene of interest discovered using JGI.
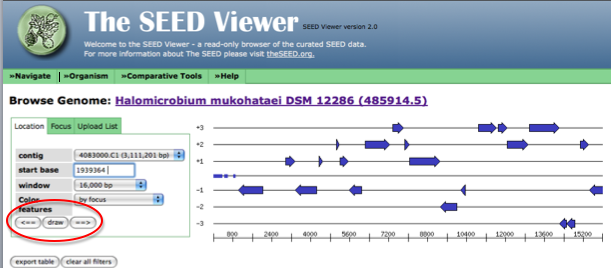
After entering the start codon, you must toggle the left and right arrows to force the genome browser to go to the location of interest in the genome. First, click on the arrow pointing right, next point on the arrow pointing left.
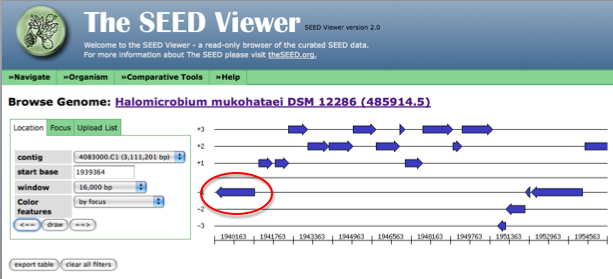
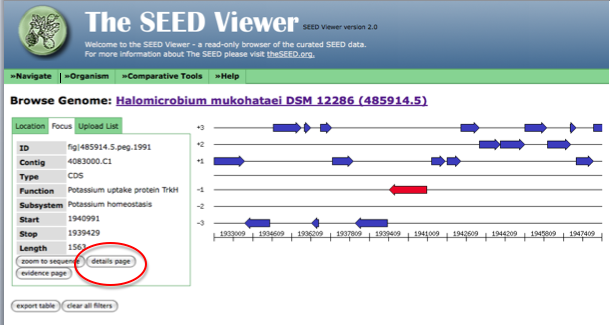
Now, the arrow in the genome diagram closest to the left of the page (if a called gene for this location exists) will be the gene sharing a start or stop codon with JGI. Click on this arrow (it will turn red) and information about this gene, including a link to the gene details page (circled in red) will appear to the left of the diagram.
The following figure shows how the page looks after clicking on the arrow representing your gene of interest. Follow the link to the details page for more information on this gene.
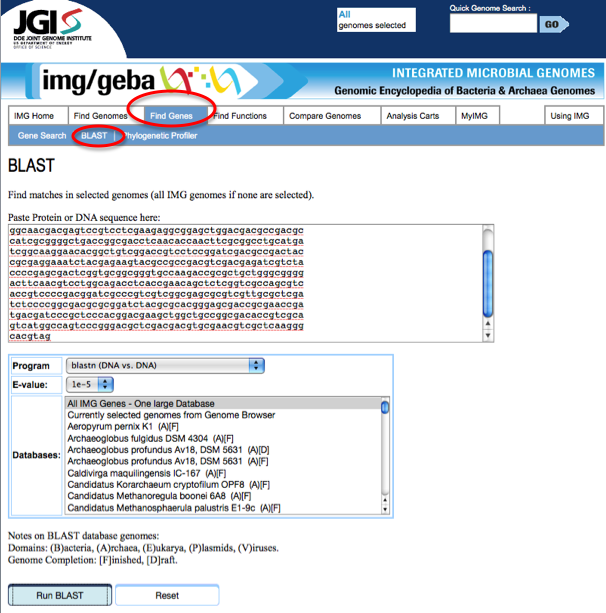
If a gene of interest is discovered in RAST and you want to find out if this gene exists in JGI, from the JGI home page, click on the Find Genes tab, then the BLAST link:
Paste the sequence of your gene of interest obtained from RAST into the field called Paste Protein or DNA sequence here:. Also, in the drop down menu labeled Program choose blastn (DNA vs DNA). Next, click run blast at the bottom of the page.
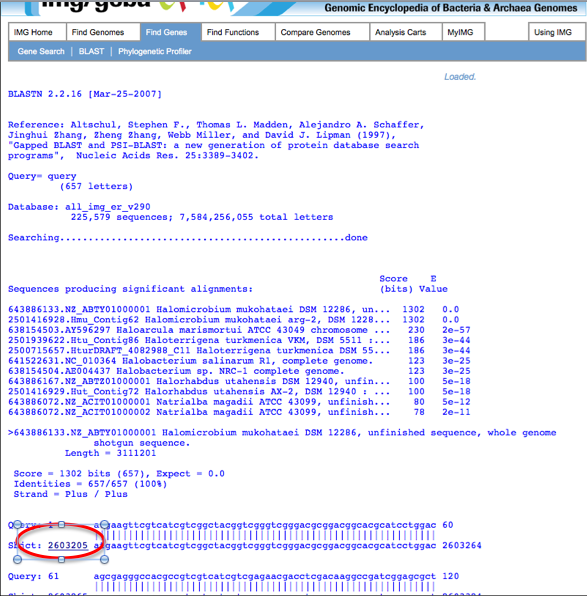
The results page will appear as the following screen shot shows:
If your gene of interest has been called by JGI, a link to the gene will appear next to the matching sequence.
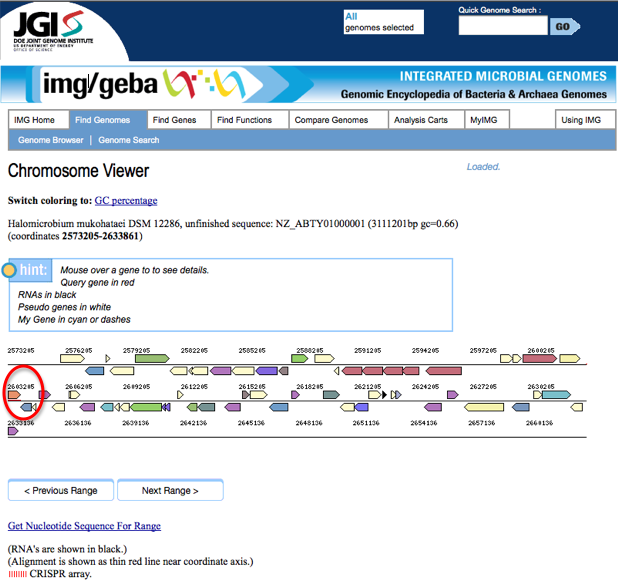
Clicking on the link will bring you to a page that looks like the following screen shot. The portion underlined in red is the gene of interest. Clicking on the arrow above this red line will bring you to an information page about the gene.