Histone Deacetylase (Katie Richeson)
Contents
Histone Deacetylase (644029844)
What is this gene?
The Adopt a Genome Project claims that this gene's functionality is as a [http://img.jgi.doe.gov/cgi-bin/geba/main.cgi?section=GeneDetail&page=geneDetail&gene_oid=644029844 deacetylase, histone deacetylase/acetoin utilization protein.
These two proteins facilitate the removal of acetyl groups from histones in eukarya and are share 9 motifs with similar genes/proteins in archaea and eubacteria [Leipe and Landsman]. This is where our species comes into play: as an archaea we should ask ourselves where this gene comes from and whether or not the normal functioning of the gene in eukaraya is also seen in our species. If so this means that our archaea has the ability to epigenetically alter transcription of specific genes. This would be an interesting study to show evolutionary patterns, as bacteria do not normally posses histone deacetylases.
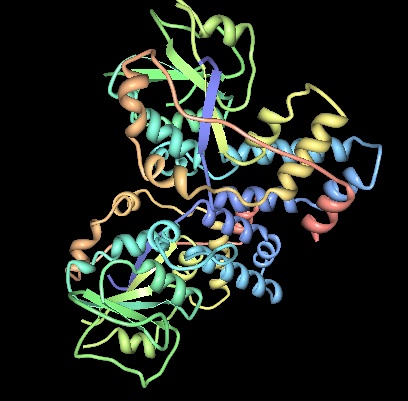
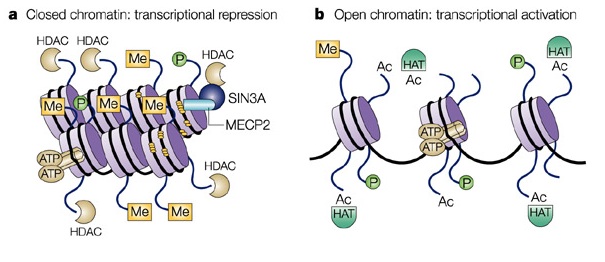
What is a Histone Deacetylase?
Histone deacetylase (HDAC) and histone acetyltransferase (HAT) are enzymes that influence transcription by removing or adding acetyl groups to lysine located near the amino termini of histone proteins. When HAT adds an acetyl group to a histone transcriptional activity changes, whereas deactylation leads to gene silencing. "HDACs are also involved in the reversible acetylation of non-histone proteins" [Vinci-Biochem].
Histone Acetylation in eukarya is a form of epigenetic regulation, where the density of chromatin is affected leading to changes in transcriptional rates.
Blastn Search
In completing a blastn search for this gene sequence I found many different hits from other halophiles but none of the hits described any sort of function. The results only showed that this sequence is often found in other halophiles, such as Haloarcula marismortui.
Blastx Search
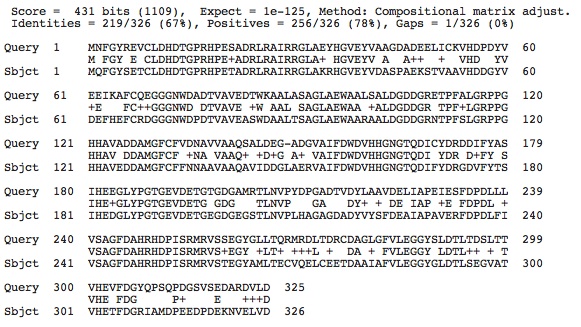
When I did a blastx search using the sequence of our specie's histone deacetylase multiple histone deacetylase protein super family alignments appeared (this is also where I got the amino acid sequence from the nucleotide sequence for this gene in our species). Below is a picture of the matches from the blastx results:
Blastp Alignment with Other Halophiles
I next went on the compare the sequence to halophile genomes that have already been annotated in order to determine if the function of similar sequences was the same. I started with Haloferax volcanii using the GenBank annotation data and found 1 histone deacetylase gene and 3 histone acetylases. I think it is important to note that while searching the list of functional protein genes for our species I found no histone acetyltransferase genes, which asks the question. . . What is placing the acetyl groups on the histone that the deacetylase is removing OR is the deacetylase really doing its job?
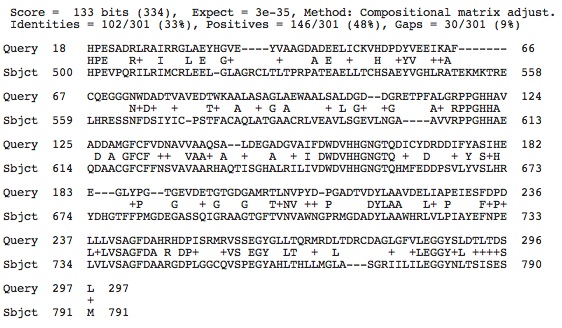
Below is the data from the blast2 results comparing the histone deacetylase protein sequences from our species and Haloferax volcanii. They are 78% the same when positives are taken into account. I would say that they are certainly similar but not extremely similar.
I did the same paralog search in the halophile Haloferax mediteranei from Spain using GenBank annotation results. This genome included one histone deacetylase and 2 histone acetyltransferases. The blastp alignment results are seen below. I would say that little of this amino acid sequence is conserved in this halophile because only a small part of the sequences match up.
Blastp Alignment with Human Histone Deacetylase Gene
I also searched the human genome to determine how many histone deacetylase genes existed and did a blastp alignment search between these two genes. I used Ensembl to look up amino acid sequences for histone deacetylases in the human genome and found a few results. The blastp results between one human histone deacetylase gene and the histone deacetylase gene in out species are as follows:
GC Content
Finally I explored the GC content of this gene to determine if it originates from another genome. The GC content of our species' genome is around 65%. The GC content for this gene is 68%. The similarity between these two GC content percentages seems to indicate that this gene is not an alien gene resulting from some sort of lateral transfer, but it does not rule out that this gene may be an ortholog.