Difference between revisions of "Davidson Missouri W/Solving the HPP in vivo"
(→Flipping Pancakes) |
Amshoecraft (talk | contribs) |
||
| (20 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | <center>[[Davidson Missouri W| <span style="color:red">Home</span>]] | [[Davidson Missouri W/Background Information| <span style="color:red">Background Information</span>]] | [[Davidson Missouri W/Solving the HPP in vivo| <span style="color:black">Current Project: Solving the Hamiltonian Path Problem ''in vivo''</span>]] | [[Davidson Missouri W/Mathematical Modeling| <span style="color:red">Mathematical Modeling</span>]] | [[Davidson Missouri W/Gene splitting| <span style="color:red"> Gene Splitting </span>]] | [[Davidson Missouri W/Controlling Expression| <span style="color:red"> Controlling Expression </span>]] | [[Davidson Missouri W/Traveling Salesperson Problem| <span style="color:red">Traveling Salesperson Problem</span> ]] | [[Davidson Missouri W/Backwards promotion and read-through transcription| <span style="color:red">Backwards Promotion and Read-Through Transcription</span>]] | [[Davidson Missouri W/Resources and Citations|<span style="color:red">Resources and Citations</span>]]</center> | |
| − | + | <hr> | |
| − | |||
| − | + | Using the Hin/''HixC'' flipping mechanism, we are developing a bacterial computer which solves a specific mathematical problem, the ''Hamiltonian Path'' problem. | |
| − | '' | ||
| − | |||
| − | |||
| − | |||
=The Hamiltonian Path Problem= | =The Hamiltonian Path Problem= | ||
| + | A Hamiltonian Path is a trip through a graph which visits each node exactly once. A graph may have multiple Hamiltonian Paths, only one, or even none. Given a graph, a starting point and an endpoint, does it contain a Hamiltonian path? | ||
| − | + | We solve our problem by transforming ''E. coli'' cells with specially engineered plasmids. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | =The | + | ==Designing a Plasmid== |
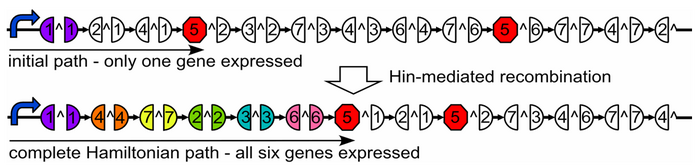
| + | Our plasmid consists of reporter genes and ''HixC'' sites. ''HixC'' sites are placed within the coding regions of our reporter genes. The reporter genes are joined in such a way as to represent a graph. Each reporter gene represents a node, and the connection of two reporter genes together without any ''HixC'' sites in between represents an edge. | ||
| − | + | [[Image:HamiltonianGraph.PNG|thumb|700px|center|Above: A graph on a plasmid. Below: flipping into a solution.]] | |
| − | == | + | ==Developing Nodes== |
| + | We represent the graph's nodes with reporter genes. In order to allow for flipping, we must insert ''HixC'' sites within the coding regions of our reporter genes. We call this process [[Gene splitting|''gene splitting'']]. If our reporter gene tolerates a ''HixC'' insertion then we can use it as a node on our graph. | ||
Latest revision as of 15:17, 9 August 2007
Using the Hin/HixC flipping mechanism, we are developing a bacterial computer which solves a specific mathematical problem, the Hamiltonian Path problem.
The Hamiltonian Path Problem
A Hamiltonian Path is a trip through a graph which visits each node exactly once. A graph may have multiple Hamiltonian Paths, only one, or even none. Given a graph, a starting point and an endpoint, does it contain a Hamiltonian path?
We solve our problem by transforming E. coli cells with specially engineered plasmids.
Designing a Plasmid
Our plasmid consists of reporter genes and HixC sites. HixC sites are placed within the coding regions of our reporter genes. The reporter genes are joined in such a way as to represent a graph. Each reporter gene represents a node, and the connection of two reporter genes together without any HixC sites in between represents an edge.
Developing Nodes
We represent the graph's nodes with reporter genes. In order to allow for flipping, we must insert HixC sites within the coding regions of our reporter genes. We call this process gene splitting. If our reporter gene tolerates a HixC insertion then we can use it as a node on our graph.