Difference between revisions of "Missing RNA genes? 2009"
Karicheson (talk | contribs) |
Karicheson (talk | contribs) |
||
| Line 1: | Line 1: | ||
In looking for other RNA genes we did the following. . . | In looking for other RNA genes we did the following. . . | ||
| − | '''1). We looked at the [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=rnas&taxon_oid=2500575004&locus_type=xRNA two "other RNA" genes ] from last year's species ([http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2500575004 Halorhabdus utahensis AX-2, DSM 12940]) and compared those sequences to [ftp://ftp.jgi-psf.org/pub/JGI_data/Microbial/halomicrobium_mukohataei_DSM_12286/080213/4083000.fsa our species entire genome ] | + | '''1). We looked at the [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=rnas&taxon_oid=2500575004&locus_type=xRNA two "other RNA" genes ] from last year's species ([http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2500575004 Halorhabdus utahensis AX-2, DSM 12940]) and compared those sequences to [ftp://ftp.jgi-psf.org/pub/JGI_data/Microbial/halomicrobium_mukohataei_DSM_12286/080213/4083000.fsa our species entire genome ] using [http://genome.jgi-psf.org/cgi-bin/runAlignment?db=halmu&advanced=1 Halomicrobium mukohataei genome portal]''' |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Results are as follows. . . | Results are as follows. . . | ||
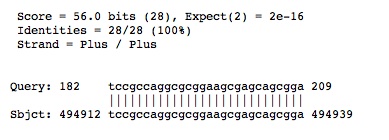
For "other RNA" gene 2500590731, named SRP_euk_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (820149..820441 (-)(293bp)): | For "other RNA" gene 2500590731, named SRP_euk_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (820149..820441 (-)(293bp)): | ||
| − | + | [http://genome.jgi-psf.org/cgi-bin/blastOutput?db=halmu&jobId=1408729&jobToken=1001272116 Genome Portal Results] | |
| − | |||
| − | |||
[[Image:portal1.JPEG]] | [[Image:portal1.JPEG]] | ||
| Line 19: | Line 12: | ||
For "other RNA" gene 2500590732, named RNaseP_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (2878153..2878553 (-)(401bp)): | For "other RNA" gene 2500590732, named RNaseP_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (2878153..2878553 (-)(401bp)): | ||
| − | + | [http://genome.jgi-psf.org/cgi-bin/blastOutput?db=halmu&jobId=1408735&jobToken=1750551800 Genome Portal Results] | |
| − | |||
| − | |||
| − | |||
[[Image:portal2.JPEG]] | [[Image:portal2.JPEG]] | ||
Revision as of 01:02, 2 September 2009
In looking for other RNA genes we did the following. . .
1). We looked at the two "other RNA" genes from last year's species (Halorhabdus utahensis AX-2, DSM 12940) and compared those sequences to our species entire genome using Halomicrobium mukohataei genome portal
Results are as follows. . .
For "other RNA" gene 2500590731, named SRP_euk_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (820149..820441 (-)(293bp)):
Genome Portal Results

For "other RNA" gene 2500590732, named RNaseP_arch, in the Halorhabdus utahensis AX-2, DSM 12940 genmone (2878153..2878553 (-)(401bp)):
Genome Portal Results

2). We also looked to ensure that all 20 amino acids had an associated tRNA gene encoded and found that all 20 amino acids were represented (although at varying frequencies).
3). We looked at the other halophiles (Halogeometricum borinquense PR3, DSM 1155 and Haliangium ochraceum SMP-2, DSM 14365) listed on the Adopt a Genome JGI site and none of them, besides the species annotated last year, had "other RNA" genes.
4). In further research we looked at other halophile genomes and used the blast from Genome Portal for all halophiles listed on the homepage. The blast for "other RNA" genes against each of the other species are as seen below. . .
A). SRP Gene
Halogeometricum borinquense
Halorhodospira halophila No hits found.
B). RNase-P Gene
Halogeometricum borinquense
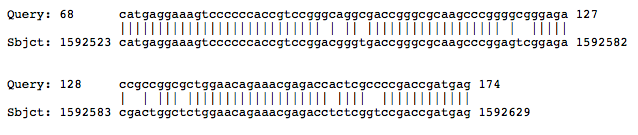 S = 109 bits (55)
E = 1e-37
Identities = 94/107 (87%)
Strand = Plus / Plus
S = 109 bits (55)
E = 1e-37
Identities = 94/107 (87%)
Strand = Plus / Plus
Halorhodospira halophila No hits found.
Halorubrum lacusprofundiS = 79.8 bits (40) E = 4e-31 Identities = 49/52 (94%) Strand = Plus / Minus
Haloterrigena turkmenicaS = 83.8 bits (42) E = 5e-26 Identities = 72/82 (87%) Strand = Plus / Plus
Halothermothrix orenii No hits found.
Halothiobacillus neapolitanus No hits found.
5). We also examined Haloferax volcanii, a species of halophile that was found through-out our blasts results while attempting to confirm annotation of tRNA genes. The genome for this halophile has previously been annotated by UCSC and researchers have not yet identified the existence of "other RNA" genes.
In the future we would like to. . .
1). Compare our species to very closely related species that have been annotated for "other RNA genes"
2). Determine what "other RNA" genes we should be looking for in archae specifically.