Difference between revisions of "Time of bloom"
| (329 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | Austin Mudd - Spring 2013 | ||
| + | <br/>Shortened URL: http://goo.gl/zuTkP | ||
| + | |||
| + | |||
__TOC__ | __TOC__ | ||
| − | === | + | ==Introduction to Flowering== |
| + | ====The Process of Flowering==== | ||
| + | *Flowering is the "switch from vegetative growth (the production of stems and leaves) to reproductive growth (the production of flowers)" ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | *The “shoot apical meristem starts to produce flowers instead of leaves” ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | *Occurs “when conditions for pollination and seed development are optimal and consequently most plants restrict flowering to a specific time of year” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | *”The genes and molecular mechanisms controlling flowering have been extensively studied in the model dicot <i>Arabidopsis thaliana</i>” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | *In <i>Arabidopsis thaliana</i>, “180 genes have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants ... Strikingly, several genes act more than once and in several tissues during floral induction” ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | |||
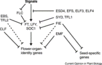
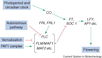
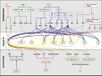
| + | [[File:Max_Planck_Institute_Pathway.png|thumb|right|alt=Diagram of pathways|The major pathways in the timing of flowering from Turck and Adrian at the [http://www.mpipz.mpg.de/305695/Project_1 Max Planck Institute for Plant Breeding Research]; permission granted]] | ||
| + | ====The Timing of Flowering==== | ||
| + | *Flowering is controlled by several “major pathways: the photoperiod and vernalization pathways control flowering in response to seasonal changes in day length and temperature; the ambient temperature pathway responds to daily growth temperatures; and the age, autonomous, and gibberellin pathways act more independently of environmental stimuli.” ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | *These “pathways converge to regulate a small number of ‘floral integrator genes,’ ... which govern flowering time by merging signals from multiple pathways” ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | |||
| + | ====The Importance of Flowering==== | ||
| + | *”Flowering is one of the most important agronomic traits influencing crop yield” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | *”Flowering time is important for adaptation to specific environments and the world's major crop species provide a particularly interesting opportunity for study because they are grown in areas outside the ecogeographical limits of their wild ancestors” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | *“Adaptation to different environments and practices has been achieved by manipulation of flowering time responses” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | *The study of flowering is ”critical for the breeding of climate change resilient crop varieties” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | *Flowering is “an excellent system for comparison between and within domestic and wild species” ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]) | ||
| + | |||
| + | |||
| + | ==Pathways Controlling Flowering== | ||
| + | ====Age Pathway==== | ||
| + | *"The miR156–SPL interaction constitutes an evolutionarily conserved, endogenous cue for both vegetative phase transition and flowering ... The age-dependent decrease in miR156 results in an increase in SPLs that promote juvenile to adult phase transition and flowering through activation of miR172, MADS box genes, and LFY" ([http://www.plantcell.org/content/early/2012/08/29/tpc.112.101014.full.pdf+html Yu ''et al.'', 2012]) | ||
| + | *5 <i>Arabidopsis</i> genes are involved in the age pathway: SPL3, SPL4, SPL5, SPL9, SPL10 ([http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2010.04148.x/pdf/ Amasino, 2010]) | ||
| + | |||
| + | ====Ambient Temperature Pathway==== | ||
| + | *Unlike "the photoperiod and vernalisation pathways [which] monitor seasonal changes in day length or temperature and ... [respond] to exposure to long days or prolonged cold temperatures, the ambient temperature pathway coordinates the response to daily growth temperatures" ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | *16 <i>Arabidopsis</i> genes are involved in the ambient temperature pathway: AGL31, ATARP6, ATBZIP27, FCA, FD, FLC, FLD, FT, FVE, MAF1, MAF3, MAF4, MAF5, SVP, TFL1, TSF ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | |||
| + | ====Autonomous Pathway==== | ||
| + | *The autonomous pathway is "activated in response to endogenous changes that are independent from the environmental cues leading to flowering", such as the plant's circadian rhythm ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | *17 <i>Arabidopsis</i> genes are involved in the autonomous pathway: CLF, FCA, FIE1, FLD, FLK, FPA, FVE, FY, LD, MSI1, SWN, VEL1, VEL2, VEL3, VIN3, VRN2, VRN5 ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | |||
| + | ====Gibberellin Pathway==== | ||
| + | *Gibberellin "is essential for floral induction in short-day conditions." In fact, plants with a "mutation in a GA biosynthetic gene, such as GA1, fail to flower" ([http://www.plantcell.org/content/early/2012/08/29/tpc.112.101014.full.pdf+html Yu ''et al.'', 2012]) | ||
| + | *5 <i>Arabidopsis</i> genes are involved in the gibberellin pathway: GAI, GID1, RGA, RGL1, RGL2 ([http://www.plantcell.org/content/early/2012/08/29/tpc.112.101014.full.pdf+html Yu ''et al.'', 2012]) | ||
| + | |||
| + | ====Light Signaling Pathway==== | ||
| + | *"Light is one of the main environmental regulators of flowering in plants. Plants sense the time of day and season of year by monitoring the light environment through light signalling pathways." Furthermore, the light signalling pathway is comprised of the "photoperiod pathway genes together with photoreceptor genes and circadian clock components" ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | *48 <i>Arabidopsis</i> genes are involved in the light signaling pathway: APRR3, APRR5, APRR9, AT1G26790, AT1G29160, AT2G34140, AT3G21320, AT3G25730, ATCOL4, ATCOL5, CCA1, CDF1, CDF2, CDF3, CDF5, CHE, CIB1, CO, COL1, COL2, COL9, COP1, CRY1, CRY2, ELF3, ELF4, ELF4-L3, FKF1, GI, LHY, LKP2, LUX, PHYA, PHYB, PHYC, PHYD, PHYE, PRR7, RAV1, RFI2, SPA1, SPA2, SPA3, SPA4, TEM1, TEM2, TOC1, ZTL ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
| + | |||
| + | ====Polycomb Pathway==== | ||
| + | *The polycomb pathway centers on “epigenetic [repression] … [of] various developmental and cellular processes … [through two] multi-subunit protein complexes: Polycomb Repressor Complex 1 (PRC1)” and Polycomb Repressor Complex 2 (PRC2) ([http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002512 Kim ''et al.'', 2012]) | ||
| + | *10 <i>Arabidopsis</i> genes are involved in the polycomb pathway: CLF, EMF1, EMF2, FIE1, FIS2, LHP1, MEA, MSI1, SWN, VRN2 ([http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002512 Kim ''et al.'', 2012]) | ||
| − | + | ====Vernalization Pathway==== | |
| − | + | *The vernalization pathway is the response to "prolonged periods of low temperature [that are required] to initiate flowering" ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | |
| + | *32 <i>Arabidopsis</i> genes are involved in the vernalization pathway: AGL14, AGL19, AGL24, AGL31, ATARP6, ATSWC6, CLF, EFS, FES1, FIE1, FLC, FRI, FRL1, FRL2, HUA2, MAF1, MAF3, MAF4, MAF5, MSI1, PAF1, PAF2, PEP, PIE1, SUF4, SVP, SWN, VEL1, VIN3, VRN1, VRN2, VRN5 ([http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]) | ||
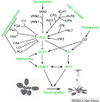
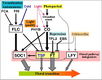
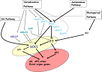
| − | == | + | ==Gallery of <i>Arabidopsis</i> Flowering Pathways== |
| − | + | <gallery widths="102px" heights="102px" perrow="6"> | |
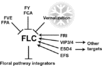
| − | + | File:Izawa_Pathway.jpeg|alt=Diagram of pathway from Izawa ''et al.'', 2003|[http://www.sciencedirect.com/science/article/pii/S1369526603000141 Izawa ''et al.'', 2003]; permission pending | |
| − | + | File:Sung_Pathway.gif|alt=Diagram of pathway from Sung ''et al.'', 2003|[http://www.sciencedirect.com/science/article/pii/S1369526602000146 Sung ''et al.'', 2003]; permission granted | |
| − | + | File:Boss_Pathway.gif|alt=Diagram of pathway from |[http://www.plantcell.org/content/16/suppl_1/S18/F2.expansion Boss ''et al.'', 2004]; permission granted | |
| − | + | File:Boss_Pathway_2.jpeg|alt=Diagram of pathway from Boss ''et al.'', 2004|[http://www.plantcell.org/content/16/suppl_1/S18/F3.expansion Boss ''et al.'', 2004]; permission granted | |
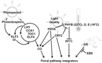
| − | + | File:Henderson_and_Dean_Pathway.jpeg|alt=Diagram of pathway from Henderson and Dean, 2004|[http://dev.biologists.org/content/131/16/3829/F1.expansion.html Henderson and Dean, 2004]; permission granted | |
| − | + | File:Amasino_Pathway.jpeg|alt=Diagram of pathway from Amasino, 2005|[http://www.sciencedirect.com/science/article/pii/S0958166905000273 Amasino, 2005]; permission granted | |
| − | + | File:He_and_Amasino_Pathway.jpeg|alt=Diagram of pathway from |[http://www.sciencedirect.com/science/article/pii/S1360138504002705 He and Amasino, 2005]; permission granted | |
| − | + | File:Yamaguchi_Pathway.jpeg|alt=Diagram of pathway from He and Amasino, 2005 |[http://pcp.oxfordjournals.org/content/46/8/1175/F7.expansion Yamaguchi ''et al.'', 2005]; permission pending | |
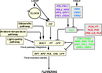
| − | + | File:Baurle_and_Dean_Pathway.jpeg|alt=Diagram of pathway from Yamaguchi ''et al.'', 2005|[http://www.sciencedirect.com/science/article/pii/S009286740600571X Bäurle and Dean, 2006]; permission granted | |
| + | File:Farrona_Pathway.jpeg|alt=Diagram of pathway from Farrona ''et al.'', 2008 |[http://www.sciencedirect.com/science/article/pii/S1084952108000566 Farrona ''et al.'', 2008]; permission granted | ||
| + | File:Farrona_Pathway_2.jpeg|alt=Diagram of pathway from |[http://www.sciencedirect.com/science/article/pii/S1084952108000566 Farrona ''et al.'', 2008]; permission granted | ||
| + | File:Amasino_Pathway_2.png|alt=Diagram of pathway from Amasino, 2010|[http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2010.04148.x/pdf/ Amasino, 2010]; permission granted | ||
| + | File:Higgins_Pathway.jpg|alt=Diagram of pathway from Higgins ''et al.'', 2010 |[http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0010065 Higgins ''et al.'', 2010]; permission granted | ||
| + | File:Kim_and_Sung_Pathway.jpeg|alt=Pathway from Kim and Sung, 2010 |[http://www.pnas.org/content/107/39/17029/F6.expansion.html Kim and Sung, 2010]; permission pending | ||
| + | File:Taiz_and_Zeiger_Pathway.jpeg|alt=Diagram of pathway from Taiz and Zeiger, 2010 |[http://5e.plantphys.net/article.php?ch=1&id=375 Taiz and Zeiger, 2010]; permission pending | ||
| + | File:Ballerini_and_Kramer_Pathway.png|alt=Diagram of pathway from Ballerini and Kramer, 2011 |[http://dash.harvard.edu/bitstream/handle/1/4903810/Ballerini%20%26%20Kramer%202011.pdf?sequence=1 Ballerini and Kramer, 2011]; permission granted | ||
| + | File:Ferrier_Pathway.jpeg|alt=Diagram of pathway from |[http://www.sciencedirect.com/science/article/pii/S0958166910002284 Ferrier ''et al.'', 2011]; permission pending | ||
| + | File:Zhang_Pathway.png|alt=Diagram of pathway from Ferrier ''et al.'', 2011|[http://www.biomedcentral.com/1471-2164/12/63 Zhang ''et al.'', 2011]; permission granted | ||
| + | File:Jung_Pathway.jpg|alt=Diagram of pathway from Jung ''et al.'', 2012 |[http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]; permission pending | ||
| + | </gallery> | ||
| + | Additional figures: | ||
| + | *[http://dev.biologists.org/content/136/20/3379/F3.expansion.html Figure 3 from Liu ''et al.'', 2009] | ||
| + | *[http://www.els.net/WileyCDA/ElsArticle/refId-a0002053.html Figure 1 from Schneitz and Balasubramanian, 2009] | ||
| + | *[http://www.sciencedirect.com/science/article/pii/S0168952510001873 Figure 1 from Wellmer and Riechmann, 2010] | ||
| + | *[http://www.sciencedirect.com/science/article/pii/S1369526611001440 Figure 1 from Posé ''et al.'', 2012] | ||
| − | |||
| − | + | ==Methods== | |
| + | ====Finding Genes==== | ||
| + | *I examined a variety of journal articles related to time of flowering in <i>Arabidopsis thaliana</i> and found a number of pathways related to flowering (see the gallery above). I came across a genomic analysis of soybean by [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012]. In this paper, they listed the "183 <i>Arabidopsis</i> genes that are known to take part in flowering regulatory pathways [taken] from previous studies." These 183 genes, plus "24 additional <i>Arabidopsis</i> genes that are grouped into the same [homolog groups] as known flowering genes," provided a solid foundation for my study. All 207 total genes from [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012] can be viewed here: [[File:Jung 207 Arabidopsis Flowering Genes.pdf]]. | ||
| + | *These 207 total genes fell into two categories: 1) flowering pathway integrators/meristem identity genes and 2) condition pathway genes (responding to the photoperiod pathway, the vernalization pathway, the ambient temperature pathway, the autonomous pathway, and other pathways). Per the direction of Dr. Jeannie Rowland of the USDA Genetic Improvement for Fruits and Vegetables Laboratory, I focused on the condition pathway genes. | ||
| + | *I identified a total of seven different pathways controlling flowering: the age pathway, the ambient temperature pathway, the autonomous pathway, the gibberellin pathway, the light signaling pathway, the polycomb pathway, and the vernalization pathway. Descriptions and primary genes involved in these pathways were taken from [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2010.04148.x/pdf/ Amasino, 2010], [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung ''et al.'', 2012], [http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1002512 Kim ''et al.'', 2012], and [http://www.plantcell.org/content/early/2012/08/29/tpc.112.101014.full.pdf+html Yu ''et al.'', 2012]. | ||
| + | *A total of 108 genes were examined, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | ====Finding SSRs==== | ||
| + | *A local database of the blueberry genome was created using the following coding: | ||
| + | <PRE>./bin/makeblastdb -in BlueberryGenome.txt -input_type fasta -dbtype nucl -title blueberry_Genome</PRE> | ||
| + | *Amino acid sequences for all <i>Arabidopsis</i> genes were taken from [http://www.arabidopsis.org/index.jsp The Arabidopsis Information Resource (TAIR) (Huala ''et al.'', 2001)]. The amino acid sequences were run via tBLASTn against the blueberry scaffolds to find the closest match using the following coding: | ||
| + | <PRE>{ echo bin/tblastn -query AASequence.txt -db BlueberryGenome.txt; bin/tblastn -query | ||
| + | AASequence.txt -db BlueberryGenome.txt; } >> AAOutput.txt</PRE> | ||
| + | *For each gene result, the best match was presumed to be the ortholog of the <i>Arabidopsis</i> gene in <i>Vaccinium corymbosum</i>. A maximum E value cutoff of e-04 was established. Although all of the results fell within this cutoff, if a tBLASTn result had not fallen below the E value limit, attempts would have been made to find and tBLASTn a <i>Vitis vinifera</i> ortholog of the <i>Arabidopsis</i> gene from [http://www.uniprot.org/uniprot/?query=organism%3A%22Vitis+vinifera+%5B29760%5D%22&sort=score UniProtKB (UniProt Consortium, 2012)] nomenclature search. | ||
| + | *SSRs were determined by importing the best match scaffold into the [http://www.vaccinium.org/cgi-bin/vaccinium_ssr SSR Tool] at the [http://www.vaccinium.org/ Genome Database for Vaccinium]. Three di/trinucleotide SSRs near the gene location on the scaffold were chosen for each gene. | ||
| − | |||
| − | + | ==SSR Results== | |
| + | All SSR results can be viewed on the [[Time of bloom SSR Results]] page. An abbreviated listing of the results is included below. | ||
| + | <br/>[[File:Flowering_SSR_Results_Table.png]] | ||
| − | {| class="wikitable" | + | |
| + | ==Flowering Genes Of Interest== | ||
| + | This table lists 108 genes involved in the age, ambient temperature, autonomous, gibberellin, light signaling, polycomb, and vernalization pathways, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." ([http://www.sciencedirect.com/science/article/pii/S0092867410004411 Fornara ''et al.'', 2010]) | ||
| + | |||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | ! <i>Arabidopsis</i> Locus !! Other Names !! AA Source !! Pathway !! width="150px" |Top Hit ''Vaccinium'' Scaffold !! E Value | ||
| + | |- | ||
| + | | AT1G01060||LATE ELONGATED HYPOCOTYL, LATE ELONGATED HYPOCOTYL 1, LHY, LHY1||[http://arabidopsis.org/servlets/TairObject?id=137165&type=locus TAIR]||Light Signaling||Scaffold00140 (length 354209) at 234299||2e-19 | ||
| + | |- | ||
| + | | AT1G02580||EMB173, EMBRYO DEFECTIVE 173, FERTILIZATION INDEPENDENT SEED 1, FIS1, MEA, MEDEA, SDG5, SET DOMAIN-CONTAINING PROTEIN 5||[http://arabidopsis.org/servlets/TairObject?id=136450&type=locus TAIR]||Polycomb||Scaffold00354 (length 215005) at 64805||4e-17 | ||
| + | |- | ||
| + | | AT1G04400||AT-PHH1, ATCRY2, CRY2, CRYPTOCHROME 2, FHA, PHH1||[http://arabidopsis.org/servlets/TairObject?id=28374&type=locus TAIR]||Light Signaling||Scaffold00649 (length 159319) at 28296||1e-119 | ||
| + | |- | ||
| + | | AT1G09570||ELONGATED HYPOCOTYL 8, FAR RED ELONGATED 1, FAR RED ELONGATED HYPOCOTYL 2, FHY2, FRE1, HY8, PHYA, PHYTOCHROME A||[http://arabidopsis.org/servlets/TairObject?id=27545&type=locus TAIR]||Light Signaling||Scaffold03861 (length 7403) at 3771||0.0 | ||
| + | |- | ||
| + | | AT1G13260||EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, RELATED TO ABI3/VP1 1||[http://arabidopsis.org/servlets/TairObject?id=137721&type=locus TAIR]||Light Signaling||Scaffold00930 (length 110378) at 63069||1e-103 | ||
| + | |- | ||
| + | | AT1G14920||GAI, GIBBERELLIC ACID INSENSITIVE, RESTORATION ON GROWTH ON AMMONIA 2, RGA2||[http://arabidopsis.org/servlets/TairObject?id=26612&type=locus TAIR]||Gibberellin||Scaffold01360 (length 81306) at 51382||0.0 | ||
| + | |- | ||
| + | | AT1G20330||COTYLEDON VASCULAR PATTERN 1, CVP1, FRILL1, FRL1, SMT2, STEROL METHYLTRANSFERASE 2||[http://arabidopsis.org/servlets/TairObject?id=27665&type=locus TAIR]||Vernalization||Scaffold02142 (length 46662) at 24743||9e-06 | ||
| + | |- | ||
| + | | AT1G22770||FB, GI, GIGANTEA||[http://arabidopsis.org/servlets/TairObject?id=136959&type=locus TAIR]||Light Signaling||Scaffold00100 (length 346620) at 198329||0.0 | ||
| + | |- | ||
| + | | AT1G25560||EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1||[http://arabidopsis.org/servlets/TairObject?id=30061&type=locus TAIR]||Light Signaling||Scaffold00930 (length 110378) at 63096||3e-99 | ||
| + | |- | ||
| + | | AT1G26790||||[http://arabidopsis.org/servlets/TairObject?id=137136&type=locus TAIR]||Light Signaling||Scaffold00079 (length 471015) at 69048||6e-33 | ||
| + | |- | ||
| + | | AT1G27370||SPL10, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 10||[http://arabidopsis.org/servlets/TairObject?id=28131&type=locus TAIR]||Age||Scaffold00127 (length 336847) at 165806||5e-33 | ||
| + | |- | ||
| + | | AT1G29160||||[http://arabidopsis.org/servlets/TairObject?id=29883&type=locus TAIR]||Light Signaling||Scaffold00270 (length 246371) at 215229||4e-50 | ||
| + | |- | ||
| + | | AT1G30970||SUF4, SUPPRESSOR OF FRIGIDA4||[http://arabidopsis.org/servlets/TairObject?id=28096&type=locus TAIR]||Vernalization||Scaffold00348 (length 210978) at 75279||1e-15 | ||
| + | |- | ||
| + | | AT1G31814||FRIGIDA LIKE 2, FRL2||[http://arabidopsis.org/servlets/TairObject?id=226970&type=locus TAIR]||Vernalization||Scaffold00289 (length 259591) at 150896||2e-19 | ||
| + | |- | ||
| + | | AT1G47250||20S PROTEASOME ALPHA SUBUNIT F2, PAF2||[http://arabidopsis.org/servlets/TairObject?id=30983&type=locus TAIR]||Vernalization||Scaffold00528 (length 197283) at 110933||3e-64 | ||
| + | |- | ||
| + | | AT1G53090||SPA1-RELATED 4, SPA4||[http://arabidopsis.org/servlets/TairObject?id=31031&type=locus TAIR]||Light Signaling||Scaffold00734 (length 158513) at 143450||3e-129 | ||
| + | |- | ||
| + | | AT1G53160||FLORAL TRANSITION AT THE MERISTEM6, FTM6, SPL4, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4||[http://arabidopsis.org/servlets/TairObject?id=27097&type=locus TAIR]||Age||Scaffold00062 (length 451336) at 412489||1e-15 | ||
| + | |- | ||
| + | | AT1G65480||FLOWERING LOCUS T, FT||[http://arabidopsis.org/servlets/TairObject?id=30541&type=locus TAIR]||Ambient Temperature||Scaffold00357 (length 260075) at 58774||6e-35 | ||
| + | |- | ||
| + | | AT1G66350||RGA-LIKE 1, RGL, RGL1||[http://arabidopsis.org/servlets/TairObject?id=137244&type=locus TAIR]||Gibberellin||Scaffold00134 (length 346346) at 174707||0.0 | ||
| + | |- | ||
| + | | AT1G68050||"FLAVIN-BINDING, KELCH REPEAT, F BOX 1", ADO3, FKF1||[http://arabidopsis.org/servlets/TairObject?id=137051&type=locus TAIR]||Light Signaling||Scaffold00110 (length 358202) at 120279||0.0 | ||
| + | |- | ||
| + | | AT1G68840||ATRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RAV2, RELATED TO ABI3/VP1 2, RELATED TO AP2 8, TEM2, TEMPRANILLO 2||[http://arabidopsis.org/servlets/TairObject?id=27570&type=locus TAIR]||Light Signaling||Scaffold00930 (length 110378) at 63096||5e-102 | ||
| + | |- | ||
| + | | AT1G77080||AGAMOUS-LIKE 27, AGL27, FLM, FLOWERING LOCUS M, MADS AFFECTING FLOWERING 1, MAF1||[http://arabidopsis.org/servlets/TairObject?id=29106&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 744||3e-13 | ||
|- | |- | ||
| − | + | | AT1G77300||ASH1 HOMOLOG 2, ASHH2, CAROTENOID CHLOROPLAST REGULATORY1, CCR1, EARLY FLOWERING IN SHORT DAYS, EFS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8||[http://arabidopsis.org/servlets/TairObject?id=136429&type=locus TAIR]||Vernalization||Scaffold00894 (length 114877) at 89213||1e-27 | |
|- | |- | ||
| − | | | + | | AT2G01570||REPRESSOR OF GA, REPRESSOR OF GA1-3 1, RGA, RGA1||[http://arabidopsis.org/servlets/TairObject?id=26549&type=locus TAIR]||Gibberellin||Scaffold01360 (length 81306) at 51382||0.0 |
|- | |- | ||
| − | | | + | | AT2G06255||ELF4-L3, ELF4-LIKE 3||[http://arabidopsis.org/servlets/TairObject?id=500231553&type=locus TAIR]||Light Signaling||Scaffold00336 (length 230445) at 188137||1e-39 |
|- | |- | ||
| − | | | + | | AT2G17770||ATBZIP27, BASIC REGION/LEUCINE ZIPPER MOTIF 27, BZIP27, FD PARALOG, FDP||[http://arabidopsis.org/servlets/TairObject?id=227228&type=locus TAIR]||Ambient Temperature||Scaffold00367 (length 240396) at 113139||7e-17 |
|- | |- | ||
| − | | | + | | AT2G18790||HY3, OOP1, OUT OF PHASE 1, PHYB, PHYTOCHROME B||[http://arabidopsis.org/servlets/TairObject?id=26548&type=locus TAIR]||Light Signaling||Scaffold00751 (length 152548) at 83698||0.0 |
|- | |- | ||
| − | | | + | | AT2G18870||VEL3, VERNALIZATION5/VIN3-LIKE 3, VIL4, VIN3-LIKE 4||[http://arabidopsis.org/servlets/TairObject?id=32249&type=locus TAIR]||Autonomous||Scaffold00396 (length 187979) at 73810||7e-19 |
|- | |- | ||
| − | | | + | | AT2G18880||VEL2, VERNALIZATION5/VIN3-LIKE 2, VIL3, VIN3-LIKE 3||[http://arabidopsis.org/servlets/TairObject?id=32230&type=locus TAIR]||Autonomous||Scaffold00026 (length 499904) at 369396||6e-41 |
|- | |- | ||
| − | | | + | | AT2G18915||ADAGIO 2, ADO2, LKP2, LOV KELCH PROTEIN 2||[http://arabidopsis.org/servlets/TairObject?id=500231567&type=locus TAIR]||Light Signaling||Scaffold00026 (length 499904) at 445582||0.0 |
|- | |- | ||
| − | | | + | | AT2G19520||ACG1, ATMSI4, FVE, MSI4, MULTICOPY SUPPRESSOR OF IRA1 4, NFC04, NFC4||[http://arabidopsis.org/servlets/TairObject?id=33019&type=locus TAIR]||Ambient Temperature, Autonomous||Scaffold00728 (length 157708) at 8716||6e-21 |
|- | |- | ||
| − | | | + | | AT2G22540||AGAMOUS-LIKE 22, AGL22, SHORT VEGETATIVE PHASE, SVP||[http://arabidopsis.org/servlets/TairObject?id=31694&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold01187 (length 101153) at 47701||2e-24 |
|- | |- | ||
| − | | | + | | AT2G23380||CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SET1, SETDOMAIN 1, SETDOMAIN GROUP 1||[http://arabidopsis.org/servlets/TairObject?id=26534&type=locus TAIR]||Autonomous, Polycomb, Vernalization||Scaffold00354 (length 215005) at 64799||5e-41 |
|- | |- | ||
| − | | | + | | AT2G25930||EARLY FLOWERING 3, ELF3, PYK20||[http://arabidopsis.org/servlets/TairObject?id=32082&type=locus TAIR]||Light Signaling||Scaffold00371 (length 223748) at 73111||6e-19 |
|- | |- | ||
| − | | | + | | AT2G32950||ARABIDOPSIS THALIANA CONSTITUTIVE PHOTOMORPHOGENIC 1, ATCOP1, CONSTITUTIVE PHOTOMORPHOGENIC 1, COP1, DEETIOLATED MUTANT 340, DET340, EMB168, EMBRYO DEFECTIVE 168, FUS1, FUSCA 1||[http://arabidopsis.org/servlets/TairObject?id=34462&type=locus TAIR]||Light Signaling||Scaffold00734 (length 158513) at 142779||5e-62 |
|- | |- | ||
| − | | | + | | AT2G33810||SPL3, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3||[http://arabidopsis.org/servlets/TairObject?id=34194&type=locus TAIR]||Age||Scaffold00062 (length 451336) at 412489||2e-17 |
|- | |- | ||
| − | | | + | | AT2G33835||FES1, FRIGIDA-ESSENTIAL 1||[http://arabidopsis.org/servlets/TairObject?id=500230872&type=locus TAIR]||Vernalization||Scaffold00102 (length 383343) at 88264||7e-17 |
|- | |- | ||
| − | | | + | | AT2G34140||||[http://arabidopsis.org/servlets/TairObject?id=33865&type=locus TAIR]||Light Signaling||Scaffold00270 (length 246371) at 215217||7e-49 |
|- | |- | ||
| − | | | + | | AT2G35670||FERTILIZATION INDEPENDENT SEED 2, FERTILIZATION-INDEPENDENT ENDOSPERM 2, FIE2, FIS2||[http://arabidopsis.org/servlets/TairObject?id=26543&type=locus TAIR]||Polycomb||Scaffold00857 (length 126644) at 55565||3e-04 |
|- | |- | ||
| − | | | + | | AT2G40080||EARLY FLOWERING 4, ELF4||[http://arabidopsis.org/servlets/TairObject?id=34766&type=locus TAIR]||Light Signaling||Scaffold00254 (length 265810) at 228805||4e-24 |
|- | |- | ||
| − | | | + | | AT2G42200||ATSPL9, SPL9, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9||[http://arabidopsis.org/servlets/TairObject?id=34557&type=locus TAIR]||Age||Scaffold00691 (length 141413) at 61795||5e-21 |
|- | |- | ||
| − | | | + | | AT2G43410||FPA||[http://arabidopsis.org/servlets/TairObject?id=34279&type=locus TAIR]||Autonomous||Scaffold01689 (length 75472) at 47663||4e-45 |
|- | |- | ||
| − | | | + | | AT2G46340||SPA1, SUPPRESSOR OF PHYA-105 1||[http://arabidopsis.org/servlets/TairObject?id=31336&type=locus TAIR]||Light Signaling||Scaffold00734 (length 158513) at 142779||3e-69 |
|- | |- | ||
| − | | | + | | AT2G46790||APRR9, ARABIDOPSIS PSEUDO-RESPONSE REGULATOR 9, PRR9, PSEUDO-RESPONSE REGULATOR 9, TL1, TOC1-LIKE PROTEIN 1||[http://arabidopsis.org/servlets/TairObject?id=32227&type=locus TAIR]||Light Signaling||Scaffold00001 (length 1030549) at 322801||1e-36 |
|- | |- | ||
| − | | | + | | AT2G46830||ATCCA1, CCA1, CIRCADIAN CLOCK ASSOCIATED 1||[http://arabidopsis.org/servlets/TairObject?id=32221&type=locus TAIR]||Light Signaling||Scaffold00140 (length 354209) at 234299||7e-20 |
|- | |- | ||
| − | | | + | | AT2G47700||RED AND FAR-RED INSENSITIVE 2, RFI2||[http://arabidopsis.org/servlets/TairObject?id=32070&type=locus TAIR]||Light Signaling||Scaffold01059 (length 101143) at 96557||1e-19 |
|- | |- | ||
| − | | | + | | AT3G02380||ATCOL2, B-BOX DOMAIN PROTEIN 3, BBX3, COL2, CONSTANS-LIKE 2||[http://arabidopsis.org/servlets/TairObject?id=35635&type=locus TAIR]||Light Signaling||Scaffold01843 (length 51900) at 40767||3e-48 |
|- | |- | ||
| − | | | + | | AT3G03450||RGA-LIKE 2, RGL2||[http://arabidopsis.org/servlets/TairObject?id=40021&type=locus TAIR]||Gibberellin||Scaffold00134 (length 346346) at 174722||0.0 |
|- | |- | ||
| − | | | + | | AT3G04610||FLK, FLOWERING LOCUS KH DOMAIN||[http://arabidopsis.org/servlets/TairObject?id=37455&type=locus TAIR]||Autonomous||Scaffold01384 (length 81068) at 28309||4e-29 |
|- | |- | ||
| − | | | + | | AT3G05120||ATGID1A, GA INSENSITIVE DWARF1A, GID1A||[http://arabidopsis.org/servlets/TairObject?id=39457&type=locus TAIR]||Gibberellin||Scaffold00101 (length 425332) at 398762||4e-175 |
|- | |- | ||
| − | | | + | | AT3G07650||B-BOX DOMAIN PROTEIN 7, BBX7, COL9, CONSTANS-LIKE 9||[http://arabidopsis.org/servlets/TairObject?id=38550&type=locus TAIR]||Light Signaling||Scaffold00832 (length 123094) at 89599||1e-55 |
|- | |- | ||
| − | | | + | | AT3G10390||FLD, FLOWERING LOCUS D||[http://arabidopsis.org/servlets/TairObject?id=35869&type=locus TAIR]||Ambient Temperature, Autonomous||Scaffold00232 (length 253418) at 139565||0.0 |
|- | |- | ||
| − | | | + | | AT3G12810||CHR13, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1, PIE1, SRCAP||[http://arabidopsis.org/servlets/TairObject?id=37964&type=locus TAIR]||Vernalization||Scaffold00147 (length 345970) at 30164||0.0 |
|- | |- | ||
| − | | | + | | AT3G15270||SPL5, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5||[http://arabidopsis.org/servlets/TairObject?id=37842&type=locus TAIR]||Age||Scaffold00105 (length 392037) at 147284||5e-16 |
|- | |- | ||
| − | | | + | | AT3G15354||SPA1-RELATED 3, SPA3||[http://arabidopsis.org/servlets/TairObject?id=500230681&type=locus TAIR]||Light Signaling||Scaffold00734 (length 158513) at 143450||1e-144 |
|- | |- | ||
| − | | | + | | AT3G18990||REDUCED VERNALIZATION RESPONSE 1, REM39, REPRODUCTIVE MERISTEM 39, VRN1||[http://arabidopsis.org/servlets/TairObject?id=37620&type=locus TAIR]||Vernalization||Scaffold00811 (length 108354) at 96661||2e-25 |
|- | |- | ||
| − | | | + | | AT3G20740||FERTILIZATION-INDEPENDENT ENDOSPERM, FERTILIZATION-INDEPENDENT ENDOSPERM 1, FIE, FIE1, FIS3||[http://arabidopsis.org/servlets/TairObject?id=38691&type=locus TAIR]||Autonomous, Polycomb, Vernalization||Scaffold01670 (length 60304) at 9836||5e-20 |
|- | |- | ||
| − | | | + | | AT3G21320||||[http://arabidopsis.org/servlets/TairObject?id=39170&type=locus TAIR]||Light Signaling||Scaffold00509 (length 192554) at 181887||1e-21 |
|- | |- | ||
| − | | | + | | AT3G24440||VERNALIZATION 5, VIL1, VIN3-LIKE 1, VRN5||[http://arabidopsis.org/servlets/TairObject?id=39196&type=locus TAIR]||Autonomous, Vernalization||Scaffold00396 (length 187979) at 73795||2e-69 |
|- | |- | ||
| − | | | + | | AT3G25730||EDF3, ETHYLENE RESPONSE DNA BINDING FACTOR 3||[http://arabidopsis.org/servlets/TairObject?id=37641&type=locus TAIR]||Light Signaling||Scaffold00930 (length 110378) at 63060||5e-100 |
|- | |- | ||
| − | | | + | | AT3G33520||ACTIN-RELATED PROTEIN 6, ARP6, ATARP6, EARLY IN SHORT DAYS 1, ESD1, SUF3, SUPPRESSOR OF FRI 3||[http://arabidopsis.org/servlets/TairObject?id=40458&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold00246 (length 286612) at 268521||1e-53 |
|- | |- | ||
| − | | | + | | AT3G46640||LUX, LUX ARRHYTHMO, PCL1, PHYTOCLOCK 1||[http://arabidopsis.org/servlets/TairObject?id=35747&type=locus TAIR]||Light Signaling||Scaffold01150 (length 88293) at 80272||3e-34 |
|- | |- | ||
| − | | | + | | AT3G47500||CDF3, CYCLING DOF FACTOR 3||[http://arabidopsis.org/servlets/TairObject?id=36439&type=locus TAIR]||Light Signaling||Scaffold00079 (length 471015) at 68175||1e-85 |
|- | |- | ||
| − | | | + | | AT4G00650||FLA, FLOWERING LOCUS A, FRI, FRIGIDA||[http://arabidopsis.org/servlets/TairObject?id=128299&type=locus TAIR]||Vernalization||Scaffold00039 (length 505275) at 216632||4e-50 |
|- | |- | ||
| − | | | + | | AT4G02020||EZA1, SDG10, SET DOMAIN-CONTAINING PROTEIN 10, SWINGER, SWN||[http://arabidopsis.org/servlets/TairObject?id=129104&type=locus TAIR]||Autonomous, Polycomb, Vernalization||Scaffold00354 (length 215005) at 64799||2e-48 |
|- | |- | ||
| − | | | + | | AT4G02560||LD, LUMINIDEPENDENS||[http://arabidopsis.org/servlets/TairObject?id=26564&type=locus TAIR]||Autonomous||Scaffold00002 (length 840149) at 158601||4e-28 |
|- | |- | ||
| − | | | + | | AT4G08920||ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, CRYPTOCHROME 1, ELONGATED HYPOCOTYL 4, HY4, OOP2, OUT OF PHASE 2||[http://arabidopsis.org/servlets/TairObject?id=129943&type=locus TAIR]||Light Signaling||Scaffold00331 (length 261439) at 124817||4e-128 |
| + | |- | ||
| + | | AT4G11110||SPA1-RELATED 2, SPA2||[http://arabidopsis.org/servlets/TairObject?id=129633&type=locus TAIR]||Light Signaling||Scaffold01034 (length 107107) at 19205||2e-62 | ||
| + | |- | ||
| + | | AT4G11880||AGAMOUS-LIKE 14, AGL14||[http://arabidopsis.org/servlets/TairObject?id=129748&type=locus TAIR]||Vernalization||Scaffold00249 (length 258199) at 131012||2e-29 | ||
| + | |- | ||
| + | | AT4G16250||PHYD, PHYTOCHROME D||[http://arabidopsis.org/servlets/TairObject?id=26567&type=locus TAIR]||Light Signaling||Scaffold00751 (length 152548) at 83707||0.0 | ||
| + | |- | ||
| + | | AT4G16280||FCA||[http://arabidopsis.org/servlets/TairObject?id=128853&type=locus TAIR]||Ambient Temperature, Autonomous||Scaffold00104 (length 360147) at 120318||5e-13 | ||
| + | |- | ||
| + | | AT4G16845||REDUCED VERNALIZATION RESPONSE 2, VRN2||[http://arabidopsis.org/servlets/TairObject?id=228106&type=locus TAIR]||Autonomous, Polycomb, Vernalization||Scaffold00857 (length 126644) at 55203||3e-07 | ||
| + | |- | ||
| + | | AT4G18130||PHYE, PHYTOCHROME E||[http://arabidopsis.org/servlets/TairObject?id=26568&type=locus TAIR]||Light Signaling||Scaffold00751 (length 152548) at 83749||0.0 | ||
| + | |- | ||
| + | | AT4G20370||TSF, TWIN SISTER OF FT||[http://arabidopsis.org/servlets/TairObject?id=26553&type=locus TAIR]||Ambient Temperature||Scaffold00357 (length 260075) at 58807||1e-33 | ||
| + | |- | ||
| + | | AT4G22950||AGAMOUS-LIKE 19, AGL19, GL19||[http://arabidopsis.org/servlets/TairObject?id=128336&type=locus TAIR]||Vernalization||Scaffold00249 (length 258199) at 131012||4e-28 | ||
| + | |- | ||
| + | | AT4G24540||AGAMOUS-LIKE 24, AGL24||[http://arabidopsis.org/servlets/TairObject?id=127586&type=locus TAIR]||Vernalization||Scaffold01187 (length 101153) at 47701||2e-25 | ||
| + | |- | ||
| + | | AT4G26000||PEP, PEPPER||[http://arabidopsis.org/servlets/TairObject?id=127403&type=locus TAIR]||Vernalization||Scaffold00021 (length 527983) at 157278||3e-29 | ||
| + | |- | ||
| + | | AT4G30200||VEL1, VERNALIZATION5/VIN3-LIKE 1, VIL2, VIN3-LIKE 2||[http://arabidopsis.org/servlets/TairObject?id=128580&type=locus TAIR]||Autonomous, Vernalization||Scaffold00026 (length 499904) at 369417||7e-64 | ||
| + | |- | ||
| + | | AT4G34530||CIB1, CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1||[http://arabidopsis.org/servlets/TairObject?id=130033&type=locus TAIR]||Light Signaling||Scaffold01322 (length 99420) at 82955||2e-29 | ||
| + | |- | ||
| + | | AT4G35900||ATBZIP14, FD, FD-1||[http://arabidopsis.org/servlets/TairObject?id=128068&type=locus TAIR]||Ambient Temperature||Scaffold00367 (length 240396) at 113139||1e-16 | ||
| + | |- | ||
| + | | AT5G02810||APRR7, PRR7, PSEUDO-RESPONSE REGULATOR 7||[http://arabidopsis.org/servlets/TairObject?id=131538&type=locus TAIR]||Light Signaling||Scaffold00125 (length 356885) at 276518||1e-33 | ||
| + | |- | ||
| + | | AT5G03840||TERMINAL FLOWER 1, TFL1||[http://arabidopsis.org/servlets/TairObject?id=131459&type=locus TAIR]||Ambient Temperature||Scaffold00181 (length 337602) at 12825||1e-28 | ||
| + | |- | ||
| + | | AT5G08330||CCA1 HIKING EXPEDITION, CHE, TRANSCRIPTION FACTOR TCP21, TCP21||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||Light Signaling||Scaffold00993 (length 109486) at 294||5e-27 | ||
| + | |- | ||
| + | | AT5G10140||AGAMOUS-LIKE 25, AGL25, FLC, FLF, FLOWERING LOCUS C, FLOWERING LOCUS F||[http://arabidopsis.org/servlets/TairObject?id=136002&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 708||8e-22 | ||
| + | |- | ||
| + | | AT5G11530||EMBRYONIC FLOWER 1, EMF1||[http://arabidopsis.org/servlets/TairObject?id=130620&type=locus TAIR]||Polycomb||Scaffold00253 (length 240879) at 11069||1e-06 | ||
| + | |- | ||
| + | | AT5G13480||FY||[http://arabidopsis.org/servlets/TairObject?id=136108&type=locus TAIR]||Autonomous||Scaffold00166 (length 328538) at 74796||2e-43 | ||
| + | |- | ||
| + | | AT5G15840||B-BOX DOMAIN PROTEIN 1, BBX1, CO, CONSTANS, FG||[http://arabidopsis.org/servlets/TairObject?id=130492&type=locus TAIR]||Light Signaling||Scaffold01843 (length 51900) at 40767||3e-29 | ||
| + | |- | ||
| + | | AT5G15850||ATCOL1, B-BOX DOMAIN PROTEIN 2, BBX2, COL1, CONSTANS-LIKE 1||[http://arabidopsis.org/servlets/TairObject?id=130495&type=locus TAIR]||Light Signaling||Scaffold01843 (length 51900) at 40767||5e-44 | ||
| + | |- | ||
| + | | AT5G17690||ATLHP1, LHP1, LIKE HETEROCHROMATIN PROTEIN 1, TERMINAL FLOWER 2, TFL2||[http://arabidopsis.org/servlets/TairObject?id=134923&type=locus TAIR]||Polycomb||Scaffold00696 (length 140617) at 128983||7e-13 | ||
| + | |- | ||
| + | | AT5G23150||ENHANCER OF AG-4 2, HUA2||[http://arabidopsis.org/servlets/TairObject?id=135220&type=locus TAIR]||Vernalization||Scaffold00686 (length 145647) at 102689||5e-42 | ||
| + | |- | ||
| + | | AT5G24470||APRR5, PRR5, PSEUDO-RESPONSE REGULATOR 5||[http://arabidopsis.org/servlets/TairObject?id=135985&type=locus TAIR]||Light Signaling||Scaffold00001 (length 1030549) at 322798||3e-40 | ||
| + | |- | ||
| + | | AT5G24930||ATCOL4, B-BOX DOMAIN PROTEIN 5, BBX5, COL4, CONSTANS-LIKE 4||[http://arabidopsis.org/servlets/TairObject?id=131285&type=locus TAIR]||Light Signaling||Scaffold11225 (length 3326) at 2816||8e-23 | ||
| + | |- | ||
| + | | AT5G35840||PHYC, PHYTOCHROME C||[http://arabidopsis.org/servlets/TairObject?id=133466&type=locus TAIR]||Light Signaling||Scaffold01070 (length 102706) at 75395||0.0 | ||
| + | |- | ||
| + | | AT5G37055||ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING||[http://arabidopsis.org/servlets/TairObject?id=500231974&type=locus TAIR]||Vernalization||Scaffold00925 (length 141061) at 38143||4e-29 | ||
| + | |- | ||
| + | | AT5G39660||CDF2, CYCLING DOF FACTOR 2||[http://arabidopsis.org/servlets/TairObject?id=133414&type=locus TAIR]||Light Signaling||Scaffold00651 (length 145047) at 19066||1e-90 | ||
| + | |- | ||
| + | | AT5G42790||ARS5, ARSENIC TOLERANCE 5, ATPSM30, PAF1, PROTEASOME ALPHA SUBUNIT F1||[http://arabidopsis.org/servlets/TairObject?id=133505&type=locus TAIR]||Vernalization||Scaffold00528 (length 197283) at 110933||5e-64 | ||
| + | |- | ||
| + | | AT5G51230||ATEMF2, CYR1, CYTOKININ RESISTANT 1, EMBRYONIC FLOWER 2, EMF2, VEF2||[http://arabidopsis.org/servlets/TairObject?id=134952&type=locus TAIR]||Polycomb||Scaffold00857 (length 126644) at 44887||9e-19 | ||
| + | |- | ||
| + | | AT5G57360||ADAGIO 1, ADO1, FKF1-LIKE PROTEIN 2, FKL2, LKP1, LOV KELCH PROTEIN 1, ZEITLUPE, ZTL||[http://arabidopsis.org/servlets/TairObject?id=134481&type=locus TAIR]||Light Signaling||Scaffold00026 (length 499904) at 445597||0.0 | ||
| + | |- | ||
| + | | AT5G57380||VERNALIZATION INSENSITIVE 3, VIN3||[http://arabidopsis.org/servlets/TairObject?id=134483&type=locus TAIR]||Autonomous, Vernalization||Scaffold00026 (length 499904) at 369405||3e-48 | ||
| + | |- | ||
| + | | AT5G57660||ATCOL5, B-BOX DOMAIN PROTEIN 6, BBX6, COL5, CONSTANS-LIKE 5||[http://arabidopsis.org/servlets/TairObject?id=134422&type=locus TAIR]||Light Signaling||Scaffold11225 (length 3326) at 2816||1e-25 | ||
| + | |- | ||
| + | | AT5G58230||ARABIDOPSIS MULTICOPY SUPRESSOR OF IRA1, ATMSI1, MATERNAL EFFECT EMBRYO ARREST 70, MEE70, MSI1, MULTICOPY SUPRESSOR OF IRA1||[http://arabidopsis.org/servlets/TairObject?id=132921&type=locus TAIR]||Autonomous, Polycomb, Vernalization||Scaffold00615 (length 179658) at 161081||2e-61 | ||
| + | |- | ||
| + | | AT5G60100||APRR3, PRR3, PSEUDO-RESPONSE REGULATOR 3||[http://arabidopsis.org/servlets/TairObject?id=133317&type=locus TAIR]||Light Signaling||Scaffold02075 (length 43325) at 14139||3e-25 | ||
| + | |- | ||
| + | | AT5G61380||APRR1, ATTOC1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TIMING OF CAB EXPRESSION 1, TOC1||[http://arabidopsis.org/servlets/TairObject?id=133196&type=locus TAIR]||Light Signaling||Scaffold00753 (length 123742) at 70468||2e-47 | ||
| + | |- | ||
| + | | AT5G62430||CDF1, CYCLING DOF FACTOR 1||[http://arabidopsis.org/servlets/TairObject?id=131911&type=locus TAIR]||Light Signaling||Scaffold01102 (length 99830) at 51435||1e-59 | ||
| + | |- | ||
| + | | AT5G65050||AGAMOUS-LIKE 31, AGL31, MADS AFFECTING FLOWERING 2, MAF2||[http://arabidopsis.org/servlets/TairObject?id=135154&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 723||4e-20 | ||
| + | |- | ||
| + | | AT5G65060||AGAMOUS-LIKE 70, AGL70, FCL3, MADS AFFECTING FLOWERING 3, MAF3||[http://arabidopsis.org/servlets/TairObject?id=130633&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 639||7e-15 | ||
| + | |- | ||
| + | | AT5G65070||AGAMOUS-LIKE 69, AGL69, FCL4, MADS AFFECTING FLOWERING 4, MAF4||[http://arabidopsis.org/servlets/TairObject?id=130634&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 723||6e-20 | ||
| + | |- | ||
| + | | AT5G65080||AGAMOUS-LIKE 68, AGL68, MADS AFFECTING FLOWERING 5, MAF5||[http://arabidopsis.org/servlets/TairObject?id=130635&type=locus TAIR]||Ambient Temperature, Vernalization||Scaffold10765 (length 2447) at 699||1e-18 | ||
|} | |} | ||
| + | |||
| + | |||
| + | ==References== | ||
| + | *Amasino RM. Seasonal and developmental timing of flowering. ''Plant J,'' '''61,''' 1001-1013 (2010). | ||
| + | *Amasino RM. Vernalization and flowering time. ''Curr Opin Plant Biol,'' '''16,''' 154–158 (2005). | ||
| + | *Ballerini ES, Kramer EM. Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia formosa. ''Evodevo,'' '''2,''' 1-20 (2011). | ||
| + | *Bäurle I, Dean C. The timing of developmental transitions in plants. ''Cell,'' '''125,''' 655-664 (2006). | ||
| + | *Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. ''Plant Cell,'' '''16,''' S18-S31 (2004). | ||
| + | *Farrona S, Coupland G, Turck F. The impact of chromatin regulation on the floral transition. ''Semin Cell Dev Biol,'' '''19,''' 560-573 (2008). | ||
| + | *Ferrier T, Matus JT, Jin J, Riechmann JL. Arabidopsis paves the way: genomic and network analyses in crops. ''Curr Opin Biotechnol,'' '''22,''' 260-270 (2011). | ||
| + | *Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. ''Cell,'' '''141,''' 550-550.e2 (2010). | ||
| + | *Henderson IR, Dean C. Control of Arabidopsis flowering: the chill before the bloom. ''Development,'' '''131,''' 3829-3838 (2004). | ||
| + | *He Y, Amasino RM. Role of chromatin modification in flowering-time control. ''Trends Plant Sci,'' '''10,''' 30-35 (2005). | ||
| + | *Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. ''PLoS One,'' '''5,''' e10065 (2010). | ||
| + | *Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. ''Nucleic Acids Res,'' '''29,''' 102-105 (2001). | ||
| + | *Izawa T, Takahashi Y, Yano M. Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. ''Curr Opin Plant Biol,'' '''6,''' 113-120 (2003). | ||
| + | *Jung CH, Wong CE, Singh MB, Bhalla PL. Comparative genomic analysis of soybean flowering genes. ''PLoS One,'' '''7,''' e38250 (2012). | ||
| + | *Kim DH, Sung S. The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. ''Proc Natl Acad Sci USA,'' '''107,''' 17029-17034 (2010). | ||
| + | *Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. ''PLoS Genet,'' '''8,''' e1002512 (2012). | ||
| + | *Liu C, Thong Z, Yu H. Coming into bloom: the specification of floral meristems. ''Development,'' '''136,''' 3379-3391 (2009). | ||
| + | *Posé D, Yant L, Schmid M. The end of innocence: flowering networks explode in complexity. ''Curr Opin Plant Biol'' '''15,''' 45-50 (2012). | ||
| + | *Schneitz K, Balasubramanian S. Floral Meristems. ''eLS'' (John Wiley & Sons Ltd, Chichester, 2009). | ||
| + | *SSR Tool. ''Genome Database for Vaccinium.'' | ||
| + | *Sung ZR, Chen L, Moon YH, Lertpiriyapong K. Mechanisms of floral repression in Arabidopsis. ''Curr Opin Plant Biol,'' '''6,''' 29-35 (2003). | ||
| + | *Taiz L, Zeiger E. ''Plant Physiology Fifth Edition Ch. 20'' (Sinauer Associates, Sunderland, MA, 2010). | ||
| + | *Turck F, Adrian J. A lesson in complexity: regulation of FLOWERING LOCUS T. ''Max Planck Institute for Plant Breeding Research.'' | ||
| + | *UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). ''Nucleic Acids Res,'' '''40,''' D71-D75 (2012). | ||
| + | *Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. ''Trends Genet,'' '''26,''' 519-527 (2010). | ||
| + | *Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. ''Plant Cell Physiol,'' '''46,''' 1175-1189 (2005). | ||
| + | *Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. ''Plant Cell,'' '''24,''' 3320-3332 (2012). | ||
| + | *Zhang JZ, Ai XY, Sun LM, Zhang DL, Guo WW, Deng XX, Hu CG. Transcriptome profile analysis of flowering molecular processes of early flowering trifoliate orange mutant and the wild-type [Poncirus trifoliata (L.) Raf.] by massively parallel signature sequencing. ''BMC Genomics,'' '''12,''' 63 (2011). | ||
Latest revision as of 16:44, 12 August 2021
Austin Mudd - Spring 2013
Shortened URL: http://goo.gl/zuTkP
Contents
Introduction to Flowering
The Process of Flowering
- Flowering is the "switch from vegetative growth (the production of stems and leaves) to reproductive growth (the production of flowers)" (Higgins et al., 2010)
- The “shoot apical meristem starts to produce flowers instead of leaves” (Fornara et al., 2010)
- Occurs “when conditions for pollination and seed development are optimal and consequently most plants restrict flowering to a specific time of year” (Higgins et al., 2010)
- ”The genes and molecular mechanisms controlling flowering have been extensively studied in the model dicot Arabidopsis thaliana” (Higgins et al., 2010)
- In Arabidopsis thaliana, “180 genes have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants ... Strikingly, several genes act more than once and in several tissues during floral induction” (Fornara et al., 2010)
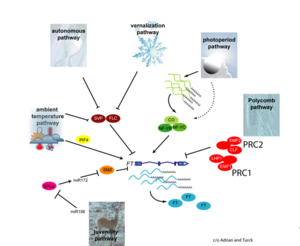
The Timing of Flowering
- Flowering is controlled by several “major pathways: the photoperiod and vernalization pathways control flowering in response to seasonal changes in day length and temperature; the ambient temperature pathway responds to daily growth temperatures; and the age, autonomous, and gibberellin pathways act more independently of environmental stimuli.” (Fornara et al., 2010)
- These “pathways converge to regulate a small number of ‘floral integrator genes,’ ... which govern flowering time by merging signals from multiple pathways” (Fornara et al., 2010)
The Importance of Flowering
- ”Flowering is one of the most important agronomic traits influencing crop yield” (Jung et al., 2012)
- ”Flowering time is important for adaptation to specific environments and the world's major crop species provide a particularly interesting opportunity for study because they are grown in areas outside the ecogeographical limits of their wild ancestors” (Higgins et al., 2010)
- “Adaptation to different environments and practices has been achieved by manipulation of flowering time responses” (Higgins et al., 2010)
- The study of flowering is ”critical for the breeding of climate change resilient crop varieties” (Jung et al., 2012)
- Flowering is “an excellent system for comparison between and within domestic and wild species” (Higgins et al., 2010)
Pathways Controlling Flowering
Age Pathway
- "The miR156–SPL interaction constitutes an evolutionarily conserved, endogenous cue for both vegetative phase transition and flowering ... The age-dependent decrease in miR156 results in an increase in SPLs that promote juvenile to adult phase transition and flowering through activation of miR172, MADS box genes, and LFY" (Yu et al., 2012)
- 5 Arabidopsis genes are involved in the age pathway: SPL3, SPL4, SPL5, SPL9, SPL10 (Amasino, 2010)
Ambient Temperature Pathway
- Unlike "the photoperiod and vernalisation pathways [which] monitor seasonal changes in day length or temperature and ... [respond] to exposure to long days or prolonged cold temperatures, the ambient temperature pathway coordinates the response to daily growth temperatures" (Jung et al., 2012)
- 16 Arabidopsis genes are involved in the ambient temperature pathway: AGL31, ATARP6, ATBZIP27, FCA, FD, FLC, FLD, FT, FVE, MAF1, MAF3, MAF4, MAF5, SVP, TFL1, TSF (Jung et al., 2012)
Autonomous Pathway
- The autonomous pathway is "activated in response to endogenous changes that are independent from the environmental cues leading to flowering", such as the plant's circadian rhythm (Jung et al., 2012)
- 17 Arabidopsis genes are involved in the autonomous pathway: CLF, FCA, FIE1, FLD, FLK, FPA, FVE, FY, LD, MSI1, SWN, VEL1, VEL2, VEL3, VIN3, VRN2, VRN5 (Jung et al., 2012)
Gibberellin Pathway
- Gibberellin "is essential for floral induction in short-day conditions." In fact, plants with a "mutation in a GA biosynthetic gene, such as GA1, fail to flower" (Yu et al., 2012)
- 5 Arabidopsis genes are involved in the gibberellin pathway: GAI, GID1, RGA, RGL1, RGL2 (Yu et al., 2012)
Light Signaling Pathway
- "Light is one of the main environmental regulators of flowering in plants. Plants sense the time of day and season of year by monitoring the light environment through light signalling pathways." Furthermore, the light signalling pathway is comprised of the "photoperiod pathway genes together with photoreceptor genes and circadian clock components" (Jung et al., 2012)
- 48 Arabidopsis genes are involved in the light signaling pathway: APRR3, APRR5, APRR9, AT1G26790, AT1G29160, AT2G34140, AT3G21320, AT3G25730, ATCOL4, ATCOL5, CCA1, CDF1, CDF2, CDF3, CDF5, CHE, CIB1, CO, COL1, COL2, COL9, COP1, CRY1, CRY2, ELF3, ELF4, ELF4-L3, FKF1, GI, LHY, LKP2, LUX, PHYA, PHYB, PHYC, PHYD, PHYE, PRR7, RAV1, RFI2, SPA1, SPA2, SPA3, SPA4, TEM1, TEM2, TOC1, ZTL (Jung et al., 2012)
Polycomb Pathway
- The polycomb pathway centers on “epigenetic [repression] … [of] various developmental and cellular processes … [through two] multi-subunit protein complexes: Polycomb Repressor Complex 1 (PRC1)” and Polycomb Repressor Complex 2 (PRC2) (Kim et al., 2012)
- 10 Arabidopsis genes are involved in the polycomb pathway: CLF, EMF1, EMF2, FIE1, FIS2, LHP1, MEA, MSI1, SWN, VRN2 (Kim et al., 2012)
Vernalization Pathway
- The vernalization pathway is the response to "prolonged periods of low temperature [that are required] to initiate flowering" (Jung et al., 2012)
- 32 Arabidopsis genes are involved in the vernalization pathway: AGL14, AGL19, AGL24, AGL31, ATARP6, ATSWC6, CLF, EFS, FES1, FIE1, FLC, FRI, FRL1, FRL2, HUA2, MAF1, MAF3, MAF4, MAF5, MSI1, PAF1, PAF2, PEP, PIE1, SUF4, SVP, SWN, VEL1, VIN3, VRN1, VRN2, VRN5 (Jung et al., 2012)
Gallery of Arabidopsis Flowering Pathways
Izawa et al., 2003; permission pending
Sung et al., 2003; permission granted
Boss et al., 2004; permission granted
Boss et al., 2004; permission granted
Henderson and Dean, 2004; permission granted
Amasino, 2005; permission granted
He and Amasino, 2005; permission granted
Yamaguchi et al., 2005; permission pending
Bäurle and Dean, 2006; permission granted
Farrona et al., 2008; permission granted
Farrona et al., 2008; permission granted
Amasino, 2010; permission granted
Higgins et al., 2010; permission granted
Kim and Sung, 2010; permission pending
Taiz and Zeiger, 2010; permission pending
Ballerini and Kramer, 2011; permission granted
Ferrier et al., 2011; permission pending
Zhang et al., 2011; permission granted
Jung et al., 2012; permission pending
Additional figures:
- Figure 3 from Liu et al., 2009
- Figure 1 from Schneitz and Balasubramanian, 2009
- Figure 1 from Wellmer and Riechmann, 2010
- Figure 1 from Posé et al., 2012
Methods
Finding Genes
- I examined a variety of journal articles related to time of flowering in Arabidopsis thaliana and found a number of pathways related to flowering (see the gallery above). I came across a genomic analysis of soybean by Jung et al., 2012. In this paper, they listed the "183 Arabidopsis genes that are known to take part in flowering regulatory pathways [taken] from previous studies." These 183 genes, plus "24 additional Arabidopsis genes that are grouped into the same [homolog groups] as known flowering genes," provided a solid foundation for my study. All 207 total genes from Jung et al., 2012 can be viewed here: File:Jung 207 Arabidopsis Flowering Genes.pdf.
- These 207 total genes fell into two categories: 1) flowering pathway integrators/meristem identity genes and 2) condition pathway genes (responding to the photoperiod pathway, the vernalization pathway, the ambient temperature pathway, the autonomous pathway, and other pathways). Per the direction of Dr. Jeannie Rowland of the USDA Genetic Improvement for Fruits and Vegetables Laboratory, I focused on the condition pathway genes.
- I identified a total of seven different pathways controlling flowering: the age pathway, the ambient temperature pathway, the autonomous pathway, the gibberellin pathway, the light signaling pathway, the polycomb pathway, and the vernalization pathway. Descriptions and primary genes involved in these pathways were taken from Amasino, 2010, Jung et al., 2012, Kim et al., 2012, and Yu et al., 2012.
- A total of 108 genes were examined, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." (Fornara et al., 2010)
Finding SSRs
- A local database of the blueberry genome was created using the following coding:
./bin/makeblastdb -in BlueberryGenome.txt -input_type fasta -dbtype nucl -title blueberry_Genome
- Amino acid sequences for all Arabidopsis genes were taken from The Arabidopsis Information Resource (TAIR) (Huala et al., 2001). The amino acid sequences were run via tBLASTn against the blueberry scaffolds to find the closest match using the following coding:
{ echo bin/tblastn -query AASequence.txt -db BlueberryGenome.txt; bin/tblastn -query
AASequence.txt -db BlueberryGenome.txt; } >> AAOutput.txt
- For each gene result, the best match was presumed to be the ortholog of the Arabidopsis gene in Vaccinium corymbosum. A maximum E value cutoff of e-04 was established. Although all of the results fell within this cutoff, if a tBLASTn result had not fallen below the E value limit, attempts would have been made to find and tBLASTn a Vitis vinifera ortholog of the Arabidopsis gene from UniProtKB (UniProt Consortium, 2012) nomenclature search.
- SSRs were determined by importing the best match scaffold into the SSR Tool at the Genome Database for Vaccinium. Three di/trinucleotide SSRs near the gene location on the scaffold were chosen for each gene.
SSR Results
All SSR results can be viewed on the Time of bloom SSR Results page. An abbreviated listing of the results is included below.
Flowering Genes Of Interest
This table lists 108 genes involved in the age, ambient temperature, autonomous, gibberellin, light signaling, polycomb, and vernalization pathways, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." (Fornara et al., 2010)
| Arabidopsis Locus | Other Names | AA Source | Pathway | Top Hit Vaccinium Scaffold | E Value |
|---|---|---|---|---|---|
| AT1G01060 | LATE ELONGATED HYPOCOTYL, LATE ELONGATED HYPOCOTYL 1, LHY, LHY1 | TAIR | Light Signaling | Scaffold00140 (length 354209) at 234299 | 2e-19 |
| AT1G02580 | EMB173, EMBRYO DEFECTIVE 173, FERTILIZATION INDEPENDENT SEED 1, FIS1, MEA, MEDEA, SDG5, SET DOMAIN-CONTAINING PROTEIN 5 | TAIR | Polycomb | Scaffold00354 (length 215005) at 64805 | 4e-17 |
| AT1G04400 | AT-PHH1, ATCRY2, CRY2, CRYPTOCHROME 2, FHA, PHH1 | TAIR | Light Signaling | Scaffold00649 (length 159319) at 28296 | 1e-119 |
| AT1G09570 | ELONGATED HYPOCOTYL 8, FAR RED ELONGATED 1, FAR RED ELONGATED HYPOCOTYL 2, FHY2, FRE1, HY8, PHYA, PHYTOCHROME A | TAIR | Light Signaling | Scaffold03861 (length 7403) at 3771 | 0.0 |
| AT1G13260 | EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, RELATED TO ABI3/VP1 1 | TAIR | Light Signaling | Scaffold00930 (length 110378) at 63069 | 1e-103 |
| AT1G14920 | GAI, GIBBERELLIC ACID INSENSITIVE, RESTORATION ON GROWTH ON AMMONIA 2, RGA2 | TAIR | Gibberellin | Scaffold01360 (length 81306) at 51382 | 0.0 |
| AT1G20330 | COTYLEDON VASCULAR PATTERN 1, CVP1, FRILL1, FRL1, SMT2, STEROL METHYLTRANSFERASE 2 | TAIR | Vernalization | Scaffold02142 (length 46662) at 24743 | 9e-06 |
| AT1G22770 | FB, GI, GIGANTEA | TAIR | Light Signaling | Scaffold00100 (length 346620) at 198329 | 0.0 |
| AT1G25560 | EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 | TAIR | Light Signaling | Scaffold00930 (length 110378) at 63096 | 3e-99 |
| AT1G26790 | TAIR | Light Signaling | Scaffold00079 (length 471015) at 69048 | 6e-33 | |
| AT1G27370 | SPL10, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 10 | TAIR | Age | Scaffold00127 (length 336847) at 165806 | 5e-33 |
| AT1G29160 | TAIR | Light Signaling | Scaffold00270 (length 246371) at 215229 | 4e-50 | |
| AT1G30970 | SUF4, SUPPRESSOR OF FRIGIDA4 | TAIR | Vernalization | Scaffold00348 (length 210978) at 75279 | 1e-15 |
| AT1G31814 | FRIGIDA LIKE 2, FRL2 | TAIR | Vernalization | Scaffold00289 (length 259591) at 150896 | 2e-19 |
| AT1G47250 | 20S PROTEASOME ALPHA SUBUNIT F2, PAF2 | TAIR | Vernalization | Scaffold00528 (length 197283) at 110933 | 3e-64 |
| AT1G53090 | SPA1-RELATED 4, SPA4 | TAIR | Light Signaling | Scaffold00734 (length 158513) at 143450 | 3e-129 |
| AT1G53160 | FLORAL TRANSITION AT THE MERISTEM6, FTM6, SPL4, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4 | TAIR | Age | Scaffold00062 (length 451336) at 412489 | 1e-15 |
| AT1G65480 | FLOWERING LOCUS T, FT | TAIR | Ambient Temperature | Scaffold00357 (length 260075) at 58774 | 6e-35 |
| AT1G66350 | RGA-LIKE 1, RGL, RGL1 | TAIR | Gibberellin | Scaffold00134 (length 346346) at 174707 | 0.0 |
| AT1G68050 | "FLAVIN-BINDING, KELCH REPEAT, F BOX 1", ADO3, FKF1 | TAIR | Light Signaling | Scaffold00110 (length 358202) at 120279 | 0.0 |
| AT1G68840 | ATRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RAV2, RELATED TO ABI3/VP1 2, RELATED TO AP2 8, TEM2, TEMPRANILLO 2 | TAIR | Light Signaling | Scaffold00930 (length 110378) at 63096 | 5e-102 |
| AT1G77080 | AGAMOUS-LIKE 27, AGL27, FLM, FLOWERING LOCUS M, MADS AFFECTING FLOWERING 1, MAF1 | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 744 | 3e-13 |
| AT1G77300 | ASH1 HOMOLOG 2, ASHH2, CAROTENOID CHLOROPLAST REGULATORY1, CCR1, EARLY FLOWERING IN SHORT DAYS, EFS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 | TAIR | Vernalization | Scaffold00894 (length 114877) at 89213 | 1e-27 |
| AT2G01570 | REPRESSOR OF GA, REPRESSOR OF GA1-3 1, RGA, RGA1 | TAIR | Gibberellin | Scaffold01360 (length 81306) at 51382 | 0.0 |
| AT2G06255 | ELF4-L3, ELF4-LIKE 3 | TAIR | Light Signaling | Scaffold00336 (length 230445) at 188137 | 1e-39 |
| AT2G17770 | ATBZIP27, BASIC REGION/LEUCINE ZIPPER MOTIF 27, BZIP27, FD PARALOG, FDP | TAIR | Ambient Temperature | Scaffold00367 (length 240396) at 113139 | 7e-17 |
| AT2G18790 | HY3, OOP1, OUT OF PHASE 1, PHYB, PHYTOCHROME B | TAIR | Light Signaling | Scaffold00751 (length 152548) at 83698 | 0.0 |
| AT2G18870 | VEL3, VERNALIZATION5/VIN3-LIKE 3, VIL4, VIN3-LIKE 4 | TAIR | Autonomous | Scaffold00396 (length 187979) at 73810 | 7e-19 |
| AT2G18880 | VEL2, VERNALIZATION5/VIN3-LIKE 2, VIL3, VIN3-LIKE 3 | TAIR | Autonomous | Scaffold00026 (length 499904) at 369396 | 6e-41 |
| AT2G18915 | ADAGIO 2, ADO2, LKP2, LOV KELCH PROTEIN 2 | TAIR | Light Signaling | Scaffold00026 (length 499904) at 445582 | 0.0 |
| AT2G19520 | ACG1, ATMSI4, FVE, MSI4, MULTICOPY SUPPRESSOR OF IRA1 4, NFC04, NFC4 | TAIR | Ambient Temperature, Autonomous | Scaffold00728 (length 157708) at 8716 | 6e-21 |
| AT2G22540 | AGAMOUS-LIKE 22, AGL22, SHORT VEGETATIVE PHASE, SVP | TAIR | Ambient Temperature, Vernalization | Scaffold01187 (length 101153) at 47701 | 2e-24 |
| AT2G23380 | CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SET1, SETDOMAIN 1, SETDOMAIN GROUP 1 | TAIR | Autonomous, Polycomb, Vernalization | Scaffold00354 (length 215005) at 64799 | 5e-41 |
| AT2G25930 | EARLY FLOWERING 3, ELF3, PYK20 | TAIR | Light Signaling | Scaffold00371 (length 223748) at 73111 | 6e-19 |
| AT2G32950 | ARABIDOPSIS THALIANA CONSTITUTIVE PHOTOMORPHOGENIC 1, ATCOP1, CONSTITUTIVE PHOTOMORPHOGENIC 1, COP1, DEETIOLATED MUTANT 340, DET340, EMB168, EMBRYO DEFECTIVE 168, FUS1, FUSCA 1 | TAIR | Light Signaling | Scaffold00734 (length 158513) at 142779 | 5e-62 |
| AT2G33810 | SPL3, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 | TAIR | Age | Scaffold00062 (length 451336) at 412489 | 2e-17 |
| AT2G33835 | FES1, FRIGIDA-ESSENTIAL 1 | TAIR | Vernalization | Scaffold00102 (length 383343) at 88264 | 7e-17 |
| AT2G34140 | TAIR | Light Signaling | Scaffold00270 (length 246371) at 215217 | 7e-49 | |
| AT2G35670 | FERTILIZATION INDEPENDENT SEED 2, FERTILIZATION-INDEPENDENT ENDOSPERM 2, FIE2, FIS2 | TAIR | Polycomb | Scaffold00857 (length 126644) at 55565 | 3e-04 |
| AT2G40080 | EARLY FLOWERING 4, ELF4 | TAIR | Light Signaling | Scaffold00254 (length 265810) at 228805 | 4e-24 |
| AT2G42200 | ATSPL9, SPL9, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 | TAIR | Age | Scaffold00691 (length 141413) at 61795 | 5e-21 |
| AT2G43410 | FPA | TAIR | Autonomous | Scaffold01689 (length 75472) at 47663 | 4e-45 |
| AT2G46340 | SPA1, SUPPRESSOR OF PHYA-105 1 | TAIR | Light Signaling | Scaffold00734 (length 158513) at 142779 | 3e-69 |
| AT2G46790 | APRR9, ARABIDOPSIS PSEUDO-RESPONSE REGULATOR 9, PRR9, PSEUDO-RESPONSE REGULATOR 9, TL1, TOC1-LIKE PROTEIN 1 | TAIR | Light Signaling | Scaffold00001 (length 1030549) at 322801 | 1e-36 |
| AT2G46830 | ATCCA1, CCA1, CIRCADIAN CLOCK ASSOCIATED 1 | TAIR | Light Signaling | Scaffold00140 (length 354209) at 234299 | 7e-20 |
| AT2G47700 | RED AND FAR-RED INSENSITIVE 2, RFI2 | TAIR | Light Signaling | Scaffold01059 (length 101143) at 96557 | 1e-19 |
| AT3G02380 | ATCOL2, B-BOX DOMAIN PROTEIN 3, BBX3, COL2, CONSTANS-LIKE 2 | TAIR | Light Signaling | Scaffold01843 (length 51900) at 40767 | 3e-48 |
| AT3G03450 | RGA-LIKE 2, RGL2 | TAIR | Gibberellin | Scaffold00134 (length 346346) at 174722 | 0.0 |
| AT3G04610 | FLK, FLOWERING LOCUS KH DOMAIN | TAIR | Autonomous | Scaffold01384 (length 81068) at 28309 | 4e-29 |
| AT3G05120 | ATGID1A, GA INSENSITIVE DWARF1A, GID1A | TAIR | Gibberellin | Scaffold00101 (length 425332) at 398762 | 4e-175 |
| AT3G07650 | B-BOX DOMAIN PROTEIN 7, BBX7, COL9, CONSTANS-LIKE 9 | TAIR | Light Signaling | Scaffold00832 (length 123094) at 89599 | 1e-55 |
| AT3G10390 | FLD, FLOWERING LOCUS D | TAIR | Ambient Temperature, Autonomous | Scaffold00232 (length 253418) at 139565 | 0.0 |
| AT3G12810 | CHR13, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1, PIE1, SRCAP | TAIR | Vernalization | Scaffold00147 (length 345970) at 30164 | 0.0 |
| AT3G15270 | SPL5, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5 | TAIR | Age | Scaffold00105 (length 392037) at 147284 | 5e-16 |
| AT3G15354 | SPA1-RELATED 3, SPA3 | TAIR | Light Signaling | Scaffold00734 (length 158513) at 143450 | 1e-144 |
| AT3G18990 | REDUCED VERNALIZATION RESPONSE 1, REM39, REPRODUCTIVE MERISTEM 39, VRN1 | TAIR | Vernalization | Scaffold00811 (length 108354) at 96661 | 2e-25 |
| AT3G20740 | FERTILIZATION-INDEPENDENT ENDOSPERM, FERTILIZATION-INDEPENDENT ENDOSPERM 1, FIE, FIE1, FIS3 | TAIR | Autonomous, Polycomb, Vernalization | Scaffold01670 (length 60304) at 9836 | 5e-20 |
| AT3G21320 | TAIR | Light Signaling | Scaffold00509 (length 192554) at 181887 | 1e-21 | |
| AT3G24440 | VERNALIZATION 5, VIL1, VIN3-LIKE 1, VRN5 | TAIR | Autonomous, Vernalization | Scaffold00396 (length 187979) at 73795 | 2e-69 |
| AT3G25730 | EDF3, ETHYLENE RESPONSE DNA BINDING FACTOR 3 | TAIR | Light Signaling | Scaffold00930 (length 110378) at 63060 | 5e-100 |
| AT3G33520 | ACTIN-RELATED PROTEIN 6, ARP6, ATARP6, EARLY IN SHORT DAYS 1, ESD1, SUF3, SUPPRESSOR OF FRI 3 | TAIR | Ambient Temperature, Vernalization | Scaffold00246 (length 286612) at 268521 | 1e-53 |
| AT3G46640 | LUX, LUX ARRHYTHMO, PCL1, PHYTOCLOCK 1 | TAIR | Light Signaling | Scaffold01150 (length 88293) at 80272 | 3e-34 |
| AT3G47500 | CDF3, CYCLING DOF FACTOR 3 | TAIR | Light Signaling | Scaffold00079 (length 471015) at 68175 | 1e-85 |
| AT4G00650 | FLA, FLOWERING LOCUS A, FRI, FRIGIDA | TAIR | Vernalization | Scaffold00039 (length 505275) at 216632 | 4e-50 |
| AT4G02020 | EZA1, SDG10, SET DOMAIN-CONTAINING PROTEIN 10, SWINGER, SWN | TAIR | Autonomous, Polycomb, Vernalization | Scaffold00354 (length 215005) at 64799 | 2e-48 |
| AT4G02560 | LD, LUMINIDEPENDENS | TAIR | Autonomous | Scaffold00002 (length 840149) at 158601 | 4e-28 |
| AT4G08920 | ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, CRYPTOCHROME 1, ELONGATED HYPOCOTYL 4, HY4, OOP2, OUT OF PHASE 2 | TAIR | Light Signaling | Scaffold00331 (length 261439) at 124817 | 4e-128 |
| AT4G11110 | SPA1-RELATED 2, SPA2 | TAIR | Light Signaling | Scaffold01034 (length 107107) at 19205 | 2e-62 |
| AT4G11880 | AGAMOUS-LIKE 14, AGL14 | TAIR | Vernalization | Scaffold00249 (length 258199) at 131012 | 2e-29 |
| AT4G16250 | PHYD, PHYTOCHROME D | TAIR | Light Signaling | Scaffold00751 (length 152548) at 83707 | 0.0 |
| AT4G16280 | FCA | TAIR | Ambient Temperature, Autonomous | Scaffold00104 (length 360147) at 120318 | 5e-13 |
| AT4G16845 | REDUCED VERNALIZATION RESPONSE 2, VRN2 | TAIR | Autonomous, Polycomb, Vernalization | Scaffold00857 (length 126644) at 55203 | 3e-07 |
| AT4G18130 | PHYE, PHYTOCHROME E | TAIR | Light Signaling | Scaffold00751 (length 152548) at 83749 | 0.0 |
| AT4G20370 | TSF, TWIN SISTER OF FT | TAIR | Ambient Temperature | Scaffold00357 (length 260075) at 58807 | 1e-33 |
| AT4G22950 | AGAMOUS-LIKE 19, AGL19, GL19 | TAIR | Vernalization | Scaffold00249 (length 258199) at 131012 | 4e-28 |
| AT4G24540 | AGAMOUS-LIKE 24, AGL24 | TAIR | Vernalization | Scaffold01187 (length 101153) at 47701 | 2e-25 |
| AT4G26000 | PEP, PEPPER | TAIR | Vernalization | Scaffold00021 (length 527983) at 157278 | 3e-29 |
| AT4G30200 | VEL1, VERNALIZATION5/VIN3-LIKE 1, VIL2, VIN3-LIKE 2 | TAIR | Autonomous, Vernalization | Scaffold00026 (length 499904) at 369417 | 7e-64 |
| AT4G34530 | CIB1, CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1 | TAIR | Light Signaling | Scaffold01322 (length 99420) at 82955 | 2e-29 |
| AT4G35900 | ATBZIP14, FD, FD-1 | TAIR | Ambient Temperature | Scaffold00367 (length 240396) at 113139 | 1e-16 |
| AT5G02810 | APRR7, PRR7, PSEUDO-RESPONSE REGULATOR 7 | TAIR | Light Signaling | Scaffold00125 (length 356885) at 276518 | 1e-33 |
| AT5G03840 | TERMINAL FLOWER 1, TFL1 | TAIR | Ambient Temperature | Scaffold00181 (length 337602) at 12825 | 1e-28 |
| AT5G08330 | CCA1 HIKING EXPEDITION, CHE, TRANSCRIPTION FACTOR TCP21, TCP21 | UniProtKB | Light Signaling | Scaffold00993 (length 109486) at 294 | 5e-27 |
| AT5G10140 | AGAMOUS-LIKE 25, AGL25, FLC, FLF, FLOWERING LOCUS C, FLOWERING LOCUS F | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 708 | 8e-22 |
| AT5G11530 | EMBRYONIC FLOWER 1, EMF1 | TAIR | Polycomb | Scaffold00253 (length 240879) at 11069 | 1e-06 |
| AT5G13480 | FY | TAIR | Autonomous | Scaffold00166 (length 328538) at 74796 | 2e-43 |
| AT5G15840 | B-BOX DOMAIN PROTEIN 1, BBX1, CO, CONSTANS, FG | TAIR | Light Signaling | Scaffold01843 (length 51900) at 40767 | 3e-29 |
| AT5G15850 | ATCOL1, B-BOX DOMAIN PROTEIN 2, BBX2, COL1, CONSTANS-LIKE 1 | TAIR | Light Signaling | Scaffold01843 (length 51900) at 40767 | 5e-44 |
| AT5G17690 | ATLHP1, LHP1, LIKE HETEROCHROMATIN PROTEIN 1, TERMINAL FLOWER 2, TFL2 | TAIR | Polycomb | Scaffold00696 (length 140617) at 128983 | 7e-13 |
| AT5G23150 | ENHANCER OF AG-4 2, HUA2 | TAIR | Vernalization | Scaffold00686 (length 145647) at 102689 | 5e-42 |
| AT5G24470 | APRR5, PRR5, PSEUDO-RESPONSE REGULATOR 5 | TAIR | Light Signaling | Scaffold00001 (length 1030549) at 322798 | 3e-40 |
| AT5G24930 | ATCOL4, B-BOX DOMAIN PROTEIN 5, BBX5, COL4, CONSTANS-LIKE 4 | TAIR | Light Signaling | Scaffold11225 (length 3326) at 2816 | 8e-23 |
| AT5G35840 | PHYC, PHYTOCHROME C | TAIR | Light Signaling | Scaffold01070 (length 102706) at 75395 | 0.0 |
| AT5G37055 | ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING | TAIR | Vernalization | Scaffold00925 (length 141061) at 38143 | 4e-29 |
| AT5G39660 | CDF2, CYCLING DOF FACTOR 2 | TAIR | Light Signaling | Scaffold00651 (length 145047) at 19066 | 1e-90 |
| AT5G42790 | ARS5, ARSENIC TOLERANCE 5, ATPSM30, PAF1, PROTEASOME ALPHA SUBUNIT F1 | TAIR | Vernalization | Scaffold00528 (length 197283) at 110933 | 5e-64 |
| AT5G51230 | ATEMF2, CYR1, CYTOKININ RESISTANT 1, EMBRYONIC FLOWER 2, EMF2, VEF2 | TAIR | Polycomb | Scaffold00857 (length 126644) at 44887 | 9e-19 |
| AT5G57360 | ADAGIO 1, ADO1, FKF1-LIKE PROTEIN 2, FKL2, LKP1, LOV KELCH PROTEIN 1, ZEITLUPE, ZTL | TAIR | Light Signaling | Scaffold00026 (length 499904) at 445597 | 0.0 |
| AT5G57380 | VERNALIZATION INSENSITIVE 3, VIN3 | TAIR | Autonomous, Vernalization | Scaffold00026 (length 499904) at 369405 | 3e-48 |
| AT5G57660 | ATCOL5, B-BOX DOMAIN PROTEIN 6, BBX6, COL5, CONSTANS-LIKE 5 | TAIR | Light Signaling | Scaffold11225 (length 3326) at 2816 | 1e-25 |
| AT5G58230 | ARABIDOPSIS MULTICOPY SUPRESSOR OF IRA1, ATMSI1, MATERNAL EFFECT EMBRYO ARREST 70, MEE70, MSI1, MULTICOPY SUPRESSOR OF IRA1 | TAIR | Autonomous, Polycomb, Vernalization | Scaffold00615 (length 179658) at 161081 | 2e-61 |
| AT5G60100 | APRR3, PRR3, PSEUDO-RESPONSE REGULATOR 3 | TAIR | Light Signaling | Scaffold02075 (length 43325) at 14139 | 3e-25 |
| AT5G61380 | APRR1, ATTOC1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TIMING OF CAB EXPRESSION 1, TOC1 | TAIR | Light Signaling | Scaffold00753 (length 123742) at 70468 | 2e-47 |
| AT5G62430 | CDF1, CYCLING DOF FACTOR 1 | TAIR | Light Signaling | Scaffold01102 (length 99830) at 51435 | 1e-59 |
| AT5G65050 | AGAMOUS-LIKE 31, AGL31, MADS AFFECTING FLOWERING 2, MAF2 | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 723 | 4e-20 |
| AT5G65060 | AGAMOUS-LIKE 70, AGL70, FCL3, MADS AFFECTING FLOWERING 3, MAF3 | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 639 | 7e-15 |
| AT5G65070 | AGAMOUS-LIKE 69, AGL69, FCL4, MADS AFFECTING FLOWERING 4, MAF4 | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 723 | 6e-20 |
| AT5G65080 | AGAMOUS-LIKE 68, AGL68, MADS AFFECTING FLOWERING 5, MAF5 | TAIR | Ambient Temperature, Vernalization | Scaffold10765 (length 2447) at 699 | 1e-18 |
References
- Amasino RM. Seasonal and developmental timing of flowering. Plant J, 61, 1001-1013 (2010).
- Amasino RM. Vernalization and flowering time. Curr Opin Plant Biol, 16, 154–158 (2005).
- Ballerini ES, Kramer EM. Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia formosa. Evodevo, 2, 1-20 (2011).
- Bäurle I, Dean C. The timing of developmental transitions in plants. Cell, 125, 655-664 (2006).
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell, 16, S18-S31 (2004).
- Farrona S, Coupland G, Turck F. The impact of chromatin regulation on the floral transition. Semin Cell Dev Biol, 19, 560-573 (2008).
- Ferrier T, Matus JT, Jin J, Riechmann JL. Arabidopsis paves the way: genomic and network analyses in crops. Curr Opin Biotechnol, 22, 260-270 (2011).
- Fornara F, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis. Cell, 141, 550-550.e2 (2010).
- Henderson IR, Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development, 131, 3829-3838 (2004).
- He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci, 10, 30-35 (2005).
- Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One, 5, e10065 (2010).
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, Mueller LA, Bhattacharyya D, Bhaya D, Sobral BW, Beavis W, Meinke DW, Town CD, Somerville C, Rhee SY. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res, 29, 102-105 (2001).
- Izawa T, Takahashi Y, Yano M. Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol, 6, 113-120 (2003).
- Jung CH, Wong CE, Singh MB, Bhalla PL. Comparative genomic analysis of soybean flowering genes. PLoS One, 7, e38250 (2012).
- Kim DH, Sung S. The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci USA, 107, 17029-17034 (2010).
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet, 8, e1002512 (2012).
- Liu C, Thong Z, Yu H. Coming into bloom: the specification of floral meristems. Development, 136, 3379-3391 (2009).
- Posé D, Yant L, Schmid M. The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol 15, 45-50 (2012).
- Schneitz K, Balasubramanian S. Floral Meristems. eLS (John Wiley & Sons Ltd, Chichester, 2009).
- SSR Tool. Genome Database for Vaccinium.
- Sung ZR, Chen L, Moon YH, Lertpiriyapong K. Mechanisms of floral repression in Arabidopsis. Curr Opin Plant Biol, 6, 29-35 (2003).
- Taiz L, Zeiger E. Plant Physiology Fifth Edition Ch. 20 (Sinauer Associates, Sunderland, MA, 2010).
- Turck F, Adrian J. A lesson in complexity: regulation of FLOWERING LOCUS T. Max Planck Institute for Plant Breeding Research.
- UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res, 40, D71-D75 (2012).
- Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends Genet, 26, 519-527 (2010).
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol, 46, 1175-1189 (2005).
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell, 24, 3320-3332 (2012).
- Zhang JZ, Ai XY, Sun LM, Zhang DL, Guo WW, Deng XX, Hu CG. Transcriptome profile analysis of flowering molecular processes of early flowering trifoliate orange mutant and the wild-type [Poncirus trifoliata (L.) Raf.] by massively parallel signature sequencing. BMC Genomics, 12, 63 (2011).