Difference between revisions of "Time of bloom"
(→Arabidopsis Flowering Genes) |
(→Arabidopsis Flowering Genes) |
||
| Line 128: | Line 128: | ||
| ATC||May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro).||[http://www.uniprot.org/uniprot/Q9ZNV5 UniProtKB]||- | | ATC||May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro).||[http://www.uniprot.org/uniprot/Q9ZNV5 UniProtKB]||- | ||
|- | |- | ||
| − | | ATCOL4||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=131285 | + | | ATCOL4||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=131285 TAIR]||L |
|- | |- | ||
| − | | ATCOL5||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=134422 | + | | ATCOL5||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=134422 TAIR]||L |
|- | |- | ||
| − | | ATSWC6||Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes.||[http://arabidopsis.org/servlets/TairObject?id=500231974 | + | | ATSWC6||Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes.||[http://arabidopsis.org/servlets/TairObject?id=500231974 TAIR]||V |
|- | |- | ||
| CAL||Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, FRUITFULL and LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Seems to be partially redundant to the function of APETALA1.||[http://www.uniprot.org/uniprot/Q39375 UniProtKB]||FPI, MI | | CAL||Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, FRUITFULL and LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Seems to be partially redundant to the function of APETALA1.||[http://www.uniprot.org/uniprot/Q39375 UniProtKB]||FPI, MI | ||
| Line 148: | Line 148: | ||
| CHE||-||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||L | | CHE||-||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||L | ||
|- | |- | ||
| − | | CIB1||Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression.||[http://arabidopsis.org/servlets/TairObject?id=130033 | + | | CIB1||Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression.||[http://arabidopsis.org/servlets/TairObject?id=130033 TAIR]||L |
|- | |- | ||
| − | | CKA1||Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis.||[http://arabidopsis.org/servlets/TairObject?id=132478 | + | | CKA1||Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis.||[http://arabidopsis.org/servlets/TairObject?id=132478 TAIR]||- |
|- | |- | ||
| − | | CKA2||Encodes the casein kinase II (CK2) catalytic subunit (alpha).||[http://arabidopsis.org/servlets/TairObject?id=37139 | + | | CKA2||Encodes the casein kinase II (CK2) catalytic subunit (alpha).||[http://arabidopsis.org/servlets/TairObject?id=37139 TAIR]||- |
|- | |- | ||
| − | | CLF||Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant.||[http://arabidopsis.org/servlets/TairObject?id=26534 | + | | CLF||Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant.||[http://arabidopsis.org/servlets/TairObject?id=26534 TAIR]||Au, V |
|- | |- | ||
| − | | CO||Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1.||[http://arabidopsis.org/servlets/TairObject?id=130492 | + | | CO||Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1.||[http://arabidopsis.org/servlets/TairObject?id=130492 TAIR]||FPI, L |
|- | |- | ||
| − | | COL1||Homologous to the flowering-time gene CONSTANS.||[http://arabidopsis.org/servlets/TairObject?id=130495 | + | | COL1||Homologous to the flowering-time gene CONSTANS.||[http://arabidopsis.org/servlets/TairObject?id=130495 TAIR]||FPI, L |
|- | |- | ||
| − | | COL2||Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins||[http://arabidopsis.org/servlets/TairObject?id=35635 | + | | COL2||Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins||[http://arabidopsis.org/servlets/TairObject?id=35635 TAIR]||FPI, L |
|- | |- | ||
| − | | COL3||Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching.||[http://arabidopsis.org/servlets/TairObject?id=32704 | + | | COL3||Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching.||[http://arabidopsis.org/servlets/TairObject?id=32704 TAIR]||- |
|- | |- | ||
| − | | COL9||This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO.||[http://arabidopsis.org/servlets/TairObject?id=38550 | + | | COL9||This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO.||[http://arabidopsis.org/servlets/TairObject?id=38550 TAIR]||L |
|- | |- | ||
| − | | COP1||Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light.||[http://arabidopsis.org/servlets/TairObject?id=34462 | + | | COP1||Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light.||[http://arabidopsis.org/servlets/TairObject?id=34462 TAIR]||L |
|- | |- | ||
| − | | CRY1||Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings.||[http://arabidopsis.org/servlets/TairObject?id=129943 | + | | CRY1||Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings.||[http://arabidopsis.org/servlets/TairObject?id=129943 TAIR]||L |
|- | |- | ||
| − | | CRY2||Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus.||[http://arabidopsis.org/servlets/TairObject?id=28374 | + | | CRY2||Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus.||[http://arabidopsis.org/servlets/TairObject?id=28374 TAIR]||L |
|- | |- | ||
| − | | DPBF2||Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons.||[http://arabidopsis.org/servlets/TairObject?id=35893 | + | | DPBF2||Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons.||[http://arabidopsis.org/servlets/TairObject?id=35893 TAIR]||- |
|- | |- | ||
| − | | E12A11||Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed.||[http://arabidopsis.org/servlets/TairObject?id=136229 | + | | E12A11||Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed.||[http://arabidopsis.org/servlets/TairObject?id=136229 TAIR]||- |
|- | |- | ||
| − | | EEL||Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo.||[http://arabidopsis.org/servlets/TairObject?id=35106 | + | | EEL||Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo.||[http://arabidopsis.org/servlets/TairObject?id=35106 TAIR]||- |
|- | |- | ||
| − | | EFS||Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development.||[http://arabidopsis.org/servlets/TairObject?id=136429 | + | | EFS||Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development.||[http://arabidopsis.org/servlets/TairObject?id=136429 TAIR]||V |
|- | |- | ||
| − | | ELF3||Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions.||[http://arabidopsis.org/servlets/TairObject?id=32082 | + | | ELF3||Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions.||[http://arabidopsis.org/servlets/TairObject?id=32082 TAIR]||L |
|- | |- | ||
| − | | ELF4||Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes.||[http://arabidopsis.org/servlets/TairObject?id=34766 | + | | ELF4||Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes.||[http://arabidopsis.org/servlets/TairObject?id=34766 TAIR]||L |
|- | |- | ||
| − | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32448 | + | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32448 TAIR]||- |
|- | |- | ||
| − | | ELF4-L2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=29911 | + | | ELF4-L2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=29911 TAIR]||- |
|- | |- | ||
| − | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231553 | + | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231553 TAIR]||L |
|- | |- | ||
| − | | ELF4-L4||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231439 | + | | ELF4-L4||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231439 TAIR]||- |
|- | |- | ||
| − | | ELF5||Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC||[http://arabidopsis.org/servlets/TairObject?id=134377 | + | | ELF5||Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC||[http://arabidopsis.org/servlets/TairObject?id=134377 TAIR]||- |
|- | |- | ||
| − | | ELF6||Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=130928 | + | | ELF6||Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=130928 TAIR]||- |
|- | |- | ||
| − | | ELF9||Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay.||[http://arabidopsis.org/servlets/TairObject?id=135649 | + | | ELF9||Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay.||[http://arabidopsis.org/servlets/TairObject?id=135649 TAIR]||FPI |
|- | |- | ||
| − | | EMF1||Involved in regulating reproductive development||[http://arabidopsis.org/servlets/TairObject?id=130620 | + | | EMF1||Involved in regulating reproductive development||[http://arabidopsis.org/servlets/TairObject?id=130620 TAIR]||- |
|- | |- | ||
| − | | EMF2||Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3.||[http://arabidopsis.org/servlets/TairObject?id=134952 | + | | EMF2||Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3.||[http://arabidopsis.org/servlets/TairObject?id=134952 TAIR]||- |
|- | |- | ||
| − | | ESD4||EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development.||[http://arabidopsis.org/servlets/TairObject?id=128953 Arabidopsis | + | | ESD4||EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development.||[http://arabidopsis.org/servlets/TairObject?id=128953 TAIR]||- |
| + | |- | ||
| + | | FCA||Involved in the promotion of the transition of the vegetative meristem to reproductive development. Four forms of the protein (alpha, beta, delta and gamma) are produced by alternative splicing. Involved in RNA-mediated chromatin silencing. At one point it was believed to act as an abscisic acid receptor but the paper describing that function was retracted.||[http://arabidopsis.org/servlets/TairObject?id=128853 TAIR]|| | ||
| + | |- | ||
| + | | FD||bZIP protein required for positive regulation of flowering. Mutants are late flowering. FD interacts with FT to promote flowering.Expressed in the shoot apex in floral anlagen, then declines in floral primordia.||[http://arabidopsis.org/servlets/TairObject?id=128068 TAIR]|| | ||
| + | |- | ||
| + | | FES1||Encodes a zinc finger domain containing protein that is expressed in the shoot/root apex and vasculature, and acts with FRI to repress flowering.FES1 mutants in a Col(FRI+) background will flower early under inductive conditions.||[http://arabidopsis.org/servlets/TairObject?id=500230872 TAIR]|| | ||
| + | |- | ||
| + | | FIE1||Encodes a protein similar to the transcriptional regular of the animal Polycomb group and is involved in regulation of establishment of anterior-posterior polar axis in the endosperm and repression of flowering during vegetative phase. Mutation leads endosperm to develop in the absence of fertilization and flowers to form in seedlings and non-reproductive organs. Also exhibits maternal effect gametophytic lethal phenotype, which is suppressed by hypomethylation. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. In the ovule, the FIE transcript levels increase transiently just after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=38691 TAIR]|| | ||
| + | |- | ||
| + | | FIS2||Encodes a negative regulator of seed development in the absence of pollination. In the ovule, the FIS2 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=26543 TAIR]|| | ||
| + | |- | ||
| + | | FKF1||Encodes FKF1, a flavin-binding kelch repeat F box protein, is clock-controlled, regulates transition to flowering. Forms a complex with GI on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=137051 TAIR]|| | ||
| + | |- | ||
| + | | FLC||MADS-box protein encoded by FLOWERING LOCUS C - transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock. Expression is downregulated during cold treatment. Vernalization, FRI and the autonomous pathway all influence the state of FLC chromatin. Both maternal and paternal alleles are reset by vernalization, but their earliest activation differs in timing and location. Histone H3 trimethylation at lysine 4 and histone acetylation are associated with active FLC expression, whereas histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC repression. Expression is also repressed by two small RNAs (30- and 24-nt) complementary to the FLC sense strand 3’ to the polyA site. The small RNAs are most likely derived from an antisense transcript of FLC. Interacts with SOC1 and FT chromatin in vivo. Member of a protein complex.||[http://arabidopsis.org/servlets/TairObject?id=136002 TAIR]|| | ||
| + | |- | ||
| + | | FLD||Encodes a plant homolog of a SWIRM domain containing protein found in histone deacetylase complexes in mammals. Lesions in FLD result in hyperacetylation of histones in FLC chromatin, up-regulation of FLC expression and extremely delayed flowering. FLD plays a key role in regulating the reproductive competence of the shoot and results in different developmental phase transitions in Arabidopsis.||[http://arabidopsis.org/servlets/TairObject?id=35869 TAIR]|| | ||
| + | |- | ||
| + | | FLK||RNA binding, nucleic acid binding; positive regulation of flower development||[http://arabidopsis.org/servlets/TairObject?id=37455 TAIR]|| | ||
| + | |- | ||
| + | | FLP1*||Encodes a small protein with unknown function and is similar to flower promoting factor 1. This gene is not expressed in apical meristem after floral induction but is expressed in roots, flowers, and in low abundance, leaves.||[http://arabidopsis.org/servlets/TairObject?id=128483 TAIR]|| | ||
| + | |- | ||
| + | | FPA||FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. Double mutants with FCA have reduced fertility and single/double mutants have defects in siRNA mediated chromatin silencing.||[http://arabidopsis.org/servlets/TairObject?id=34279 TAIR]|| | ||
| + | |- | ||
| + | | FPF1||Encodes a small protein of 12.6 kDa that regulates flowering and is involved in gibberellin signalling pathway. It is expressed in apical meristems immediately after the photoperiodic induction of flowering. Genetic interactions with flowering time and floral organ identity genes suggest that this gene may be involved in modulating the competence to flower. There are two other genes similar to FPF1, FLP1 (At4g31380) and FLP2 (no locus name yet, on BAC F8F16 on chr 4). This is so far a plant-specific gene and is only found in long-day mustard, arabidopsis, and rice.||[http://arabidopsis.org/servlets/TairObject?id=131288 TAIR]|| | ||
| + | |- | ||
| + | | FRI||Encodes a major determinant of natural variation in Arabidopsis flowering time. Dominant alleles of FRI confer a vernalization requirement causing plants to overwinter vegetatively. Many early flowering accessions carry loss-of-function fri alleles .Twenty distinct haplotypes that contain non-functional FRI alleles have been identified and the distribution analyzed in over 190 accessions. The common lab strains- Col and Ler each carry loss of function mutations in FRI.||[http://arabidopsis.org/servlets/TairObject?id=128299 TAIR]|| | ||
| + | |- | ||
| + | | FRL1||Encodes a sterol-C24-methyltransferases involved in sterol biosynthesis. Mutants display altered sterol composition, serrated petals and sepals and altered cotyledon vascular patterning as well as ectopic endoreduplication. This suggests that suppression of endoreduplication is important for petal morphogenesis and that normal sterol composition is required for this suppression.||[http://arabidopsis.org/servlets/TairObject?id=27665 TAIR]|| | ||
| + | |- | ||
| + | | FRL2||Family member of FRI-related genes that is required for the winter-annual habit. ||[http://arabidopsis.org/servlets/TairObject?id=226970 TAIR]|| | ||
| + | |- | ||
| + | | FT||FT, together with LFY, promotes flowering and is antagonistic with its homologous gene, TERMINAL FLOWER1 (TFL1). FT is expressed in leaves and is induced by long day treatment. Either the FT mRNA or protein is translocated to the shoot apex where it induces its own expression. Recent data suggests that FT protein acts as a long-range signal. FT is a target of CO and acts upstream of SOC1.||[http://arabidopsis.org/servlets/TairObject?id=30541 TAIR]|| | ||
| + | |- | ||
| + | | FUL||MADS box gene negatively regulated by APETALA1||[http://arabidopsis.org/servlets/TairObject?id=134558 TAIR]|| | ||
| + | |- | ||
| + | | FVE||Controls flowering.||[http://arabidopsis.org/servlets/TairObject?id=33019 TAIR]|| | ||
| + | |- | ||
| + | | FY||Encodes a protein with similarity to yeast Pfs2p, an mRNA processing factor. Involved in regulation of flowering time; affects FCA mRNA processing. Homozygous mutants are late flowering, null alleles are embryo lethal.||[http://arabidopsis.org/servlets/TairObject?id=136108 TAIR]|| | ||
| + | |- | ||
| + | | GBF4||Encodes a basic leucine zipper G-box binding factor that can bind to G-box motifs only as heterodimers with GBF2 or GBF3. A single amino acid change can confer G-box binding as homodimers.||[http://arabidopsis.org/servlets/TairObject?id=28911 TAIR]|| | ||
| + | |- | ||
| + | | GI||Together with CONSTANTS (CO) and FLOWERING LOCUS T (FT), GIGANTEA promotes flowering under long days in a circadian clock-controlled flowering pathway. GI acts earlier than CO and FT in the pathway by increasing CO and FT mRNA abundance. Located in the nucleus. Regulates several developmental processes, including photoperiod-mediated flowering, phytochrome B signaling, circadian clock, carbohydrate metabolism, and cold stress response. The gene's transcription is controlled by the circadian clock and it is post-transcriptionally regulated by light and dark. Forms a complex with FKF1 on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=136959 TAIR]|| | ||
| + | |- | ||
| + | | GRF1||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower||[http://arabidopsis.org/servlets/TairObject?id=34438 TAIR]|| | ||
| + | |- | ||
| + | | GRF10*||Encodes a 14-3-3 protein. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=136501 TAIR]|| | ||
| + | |- | ||
| + | | GRF11*||Encodes a 14-3-3 protein. Binds H+-ATPase in response to blue light.||[http://arabidopsis.org/servlets/TairObject?id=26877 TAIR]|| | ||
| + | |- | ||
| + | | GRF12*||Encodes a 14-3-3 protein.||[http://arabidopsis.org/servlets/TairObject?id=136716 TAIR]|| | ||
| + | |- | ||
| + | | GRF2||G-box binding factor GF14 omega encoding a 14-3-3 protein||[http://arabidopsis.org/servlets/TairObject?id=30202 TAIR]|| | ||
| + | |- | ||
| + | | GRF3||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=32311 TAIR]|| | ||
| + | |- | ||
| + | | GRF4||GF14 protein phi chain member of 14-3-3 protein family. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=137475 TAIR]|| | ||
| + | |- | ||
| + | | GRF5||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=38052 TAIR]|| | ||
| + | |- | ||
| + | | GRF6||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=33231 TAIR]|| | ||
| + | |- | ||
| + | | GRF7||Encodes GF14 ν, a 14-3-3 protein isoform (14-3-3ν).||[http://arabidopsis.org/servlets/TairObject?id=36055 TAIR]|| | ||
| + | |- | ||
| + | | GRF8||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=129479 TAIR]|| | ||
| + | |- | ||
| + | | HAP3A||Encodes a transcription factor from the nuclear factor Y (NF-Y) family, AtNF-YB1. Confers drought tolerance.||[http://arabidopsis.org/servlets/TairObject?id=35360 TAIR]|| | ||
| + | |- | ||
| + | | HAP3B||-||-|| | ||
| + | |- | ||
| + | | HAP5A||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5A) and expressed in vegetative and reproductive tissues.||[http://arabidopsis.org/servlets/TairObject?id=126547 TAIR]|| | ||
| + | |- | ||
| + | | HAP5B||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5B) and expressed in vegetative and reproductive tissues||[http://arabidopsis.org/servlets/TairObject?id=27444 TAIR]|| | ||
| + | |- | ||
| + | | HAP5C||Heme activated protein (HAP5c)||[http://arabidopsis.org/servlets/TairObject?id=30861 TAIR]|| | ||
| + | |- | ||
| + | | HUA2||Putative transcription factor. Member of the floral homeotic AGAMOUS pathway.Mutations in HUA enhance the phenotype of mild ag-4 allele. Single hua mutants are early flowering and have reduced levels of FLC mRNA. Other MADS box flowering time genes such as FLM and MAF2 also appear to be regulated by HUA2. HUA2 normally activates FLC expression and enhances AG function.||[http://arabidopsis.org/servlets/TairObject?id=135220 TAIR]|| | ||
|} | |} | ||
Revision as of 19:07, 19 February 2013
Contents
To Do
Note all Arabidopsis genes in File:Jung 207 Arabidopsis Flowering Genes.pdf from Jung et al., 2012.
Look for strawberry orthologs of Arabidopsis genes, such as in Mouhu et al., 2009.
Arabidopsis Flowering Pathway (2004-Present)
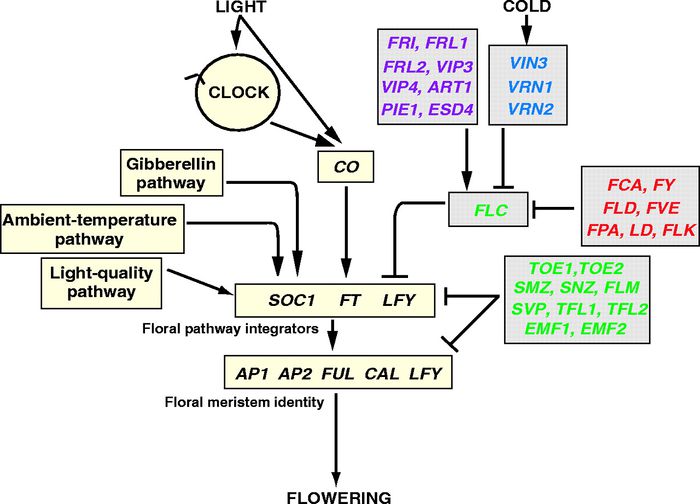
Overall pathway for the time of bloom from Henderson and Dean, 2004.
Overall pathway for the time of bloom from Schneitz, 2009.
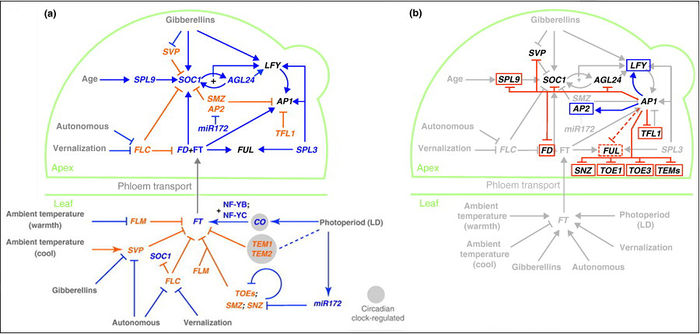
"The Arabidopsis floral transition network has been delineated as a result of extensive genetic analyses (depicted in a), but genome-wide studies have uncovered new molecular interactions and highlighted the role of AP1 as both a molecular switch and a network hub (shown in b)."
Overall pathway for the time of bloom from Wellmer and Riechmann, 2010 and Ferrier et al., 2011.
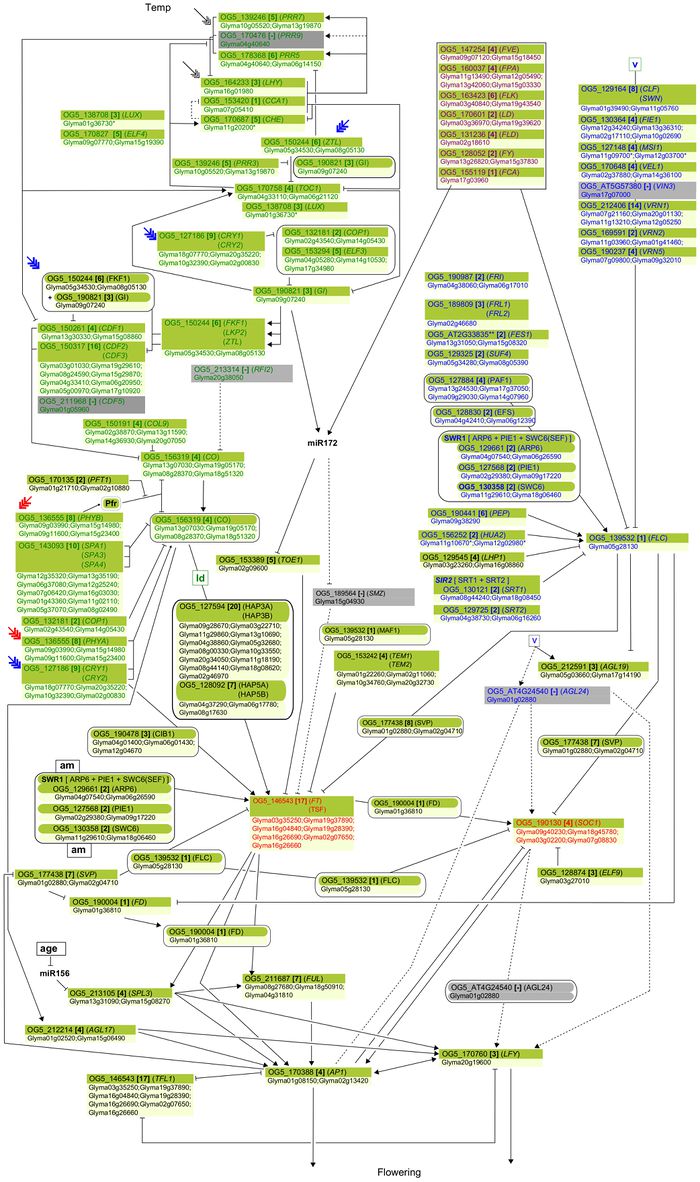
"Soybean genes marked with * are those that share the same clade with Arabidopsis genes but have not been investigated so far as flowering genes ... Arrows show promoting effects, T-bars show repressing effects. Environmental cues are shown as lower case letters in square boxes; ‘v’ is extended cold (vernalization); ‘ld’ is long days; ‘sd’ is short days; ‘am’ is ambient (non-vernalizing) temperature. Genes are shown in italics and proteins in non-italics in ovals. ‘Pfr’ indicates Pfr phytochrome signaling. Arabidopsis genes assigned to specific pathways are color-coded (photoperiod pathway in green, vernalization in blue and autonomous pathway in purple). Flowering pathway integrators are shown in red. Triple headed arrows indicate activation by red or blue light."
Overall pathway for the time of bloom from Jung et al., 2012.
All Arabidopsis Flowering Pathways
Henderson and Dean, 2004; Yamaguchi et al., 2005; Liu et al., 2009; Taiz and Zeiger, 2010; Boss et al., 2004; Zhang et al., 2011; Ballerini and Kramer, 2011; Posé et al., 2012; Kim and Sung, 2010; Wellmer and Riechmann, 2010; Farrona et al., 2008; He and Amasino, 2005; Max Planck Institute for Plant Breeding Research; Bäurle and Dean, 2006; Sung et al., 2003; Schneitz, 2009; Amasino, 2005; Ferrier et al., 2011; Izawa et al., 2003; Jung et al., 2012
Arabidopsis Flowering Genes
Note: All genes are compiled from Jung et al., 2012. Formatting is the same as Jung et al., 2012 where a * indicates 1 of "24 Arabidopsis genes, which had not been previously investigated for their roles in flowering, [that] belong to [ortholog groups] with known Arabidopsis flowering genes." Moreover, for pathways, "Am: Ambient temperature pathway, Au: Autonomous pathway, FPI: Flowering Pathway Intergrators, L: Light signaling pathway, MI: Meristem Identity, V: Vernalization pathway."
| Arabidopsis Gene | Function (All function information is directly copied from the respective AA Sequence page) | AA Sequence | Pathway |
|---|---|---|---|
| ABF1 | Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression. | UniProtKB | - |
| ABF2 | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | - |
| ABF3 | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. | UniProtKB | - |
| ABF4 | Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | - |
| ABI5 | Participates in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth. Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter and to the ABRE of the Em1 and Em6 genes promoters. Can also trans-activate its own promoter, suggesting that it is autoregulated. Plays a role in sugar-mediated senescence. | UniProtKB | - |
| AG | Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development. | UniProtKB | - |
| AGL14 | Probable transcription factor. | UniProtKB | V |
| AGL15 | Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro. | UniProtKB | - |
| AGL17 | Probable transcription factor. | UniProtKB | - |
| AGL18 | Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation. | UniProtKB | - |
| AGL19 | Probable transcription factor that promotes flowering, especially in response to vernalization by short periods of cold, in an FLC-inpedendent manner. | UniProtKB | V |
| AGL21* | Probable transcription factor. | UniProtKB | - |
| AGL24 | Transcription activator that mediates floral transition in response to vernalization. Promotes inflorescence fate in apical meristems. Acts in a dosage-dependent manner. Probably involved in the transduction of RLK-mediated signaling (e.g. IMK3 pathway). Together with AP1 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. When associated with SOC1, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. Confers inflorescence characteristics to floral primordia and early flowering. | UniProtKB | V |
| AGL31 | Probable transcription factor that prevents vernalization by short periods of cold. Acts as a floral repressor. | UniProtKB | Am, FPI, V |
| AGL6 | Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development. | UniProtKB | - |
| AGL79 | - | UniProtKB | - |
| AP1 | Transcription factor that promotes early floral meristem identity in synergy with LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Is indispensable for normal development of sepals and petals in flowers. Regulates positively the B class homeotic proteins APETALA3 and PISTILLATA with the cooperation of LEAFY and UFO. Interacts with SEPALLATA3 or AP3/PI heterodimer to form complexes that could be involved in genes regulation during floral meristem development. Regulates positively AGAMOUS in cooperation with LEAFY. Displays a redundant function with CAULIFLOWER in the up-regulation of LEAFY. Together with AGL24 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Represses flowering time genes AGL24, SVP and SOC1 in emerging floral meristems. | UniProtKB | FPI, MI |
| AP2 | Probable transcriptional activator that promotes early floral meristem identity. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Plays a central role in the specification of floral identity, particularly for the normal development of sepals and petals in the wild-type flower. Acts as A class cadastral protein by repressing the C class floral homeotic gene AGAMOUS in association with other repressors like LEUNIG and SEUSS. It is also required during seed development. | UniProtKB | - |
| AP3 | Probable transcription factor involved in the genetic control of flower development. Is required for normal development of petals and stamens in the wild-type flower. Forms a heterodimer with PISTILLATA that is required for autoregulation of both AP3 and PI genes. AP3/PI heterodimer interacts with APETALA1 or SEPALLATA3 to form a ternary complex that could be responsible for the regulation of the genes involved in the flower development. AP3/PI heterodimer activates the expression of NAP. | UniProtKB | - |
| APRR3 | Controls photoperiodic flowering response. Component of the circadian clock. Controls the degradation of APRR1/TOC1 by the SCF(ZTL) complex. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| APRR5 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| APRR9 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| AREB3 | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | UniProtKB | - |
| AT1G26790 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. | UniProtKB | L |
| AT1G29160 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Acts as a negative regulator in the phytochrome-mediated light responses. Controls phyB-mediated end-of-day response and the phyA-mediated anthocyanin accumulation. | UniProtKB | L |
| AT1G50680 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | - |
| AT1G51120 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | - |
| AT2G23070 | Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins. | UniProtKB | - |
| AT2G23080 | Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins. | UniProtKB | - |
| AT2G25920 | Uncharacterized protein | UniProtKB | - |
| AT2G34140 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. | UniProtKB | L |
| AT2G46670* | Putative uncharacterized protein | UniProtKB | L |
| AT3G21320 | Uncharacterized protein | UniProtKB | L |
| AT3G25730 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | L |
| AT4G16810 | Putative uncharacterized protein | UniProtKB | - |
| AT5G08230* | Probable transcrition factor that seems to be involved in mRNA processing. | UniProtKB | V |
| AT5G10625* | Modulates the competence to flowering of apical meristems. | UniProtKB | - |
| AT5G23280* | Transcription factor | UniProtKB | L |
| AT5G27220 | Frigida-like protein | UniProtKB | - |
| AT5G42910 | Could participate in abscisic acid-regulated gene expression. | UniProtKB | - |
| AT5G44080 | Basic leucine zipper transcription factor | UniProtKB | - |
| AT5G48250* | Zinc finger protein Constans-like 10 | UniProtKB | L |
| AT5G59570* | Putative transcription factor | UniProtKB | L |
| AT5G62040 | May form complexes with phosphorylated ligands by interfering with kinases and their effectors. | UniProtKB | - |
| ATARP6 | Component of the SWR1 complex which mediates the ATP-dependent exchange of histone H2A for the H2A variant H2A.F/Z leading to transcriptional regulation of selected genes (e.g. FLC) by chromatin remodeling. Required for the activation of FLC and FLC/MAF genes expression to levels that inhibit flowering, through both histone H3 and H4 acetylation and methylation mechanisms. Involved in several developmental processes including organization of plant organs, leaves formation, flowering time repression, and fertility. Modulates photoperiod-dependent epidermal leaves cell development; promotes cell division in long days, and cell expansion/division in short days. May be involved in the regulation of pathogenesis-related proteins (PRs). | UniProtKB | Am, V |
| ATBZIP27 | Transcription factor | GenScript | Am, MI |
| ATC | May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro). | UniProtKB | - |
| ATCOL4 | Sequence-specific DNA binding transcription factor activity, zinc ion binding. | TAIR | L |
| ATCOL5 | Sequence-specific DNA binding transcription factor activity, zinc ion binding. | TAIR | L |
| ATSWC6 | Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes. | TAIR | V |
| CAL | Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, FRUITFULL and LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Seems to be partially redundant to the function of APETALA1. | UniProtKB | FPI, MI |
| CCA1 | Transcription factor involved in the circadian clock and in the phytochrome regulation. Binds to the promoter regions of APRR1/TOC1 and TCP21/CHE to repress their transcription. Binds to the promoter regions of CAB2A and CAB2B to promote their transcription. Represses both LHY and itself. | UniProtKB | L |
| CDF1 | Cell growth defect factor | UniProtKB | L |
| CDF2 | Cell growth defect factor | UniProtKB | L |
| CDF3 | Plant non-specific lipid-transfer proteins transfer phospholipids as well as galactolipids across membranes. May play a role in wax or cutin deposition in the cell walls of expanding epidermal cells and certain secretory tissues. | UniProtKB | L |
| CDF5 | Cell growth defect factor | - | L |
| CHE | - | UniProtKB | L |
| CIB1 | Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression. | TAIR | L |
| CKA1 | Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis. | TAIR | - |
| CKA2 | Encodes the casein kinase II (CK2) catalytic subunit (alpha). | TAIR | - |
| CLF | Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant. | TAIR | Au, V |
| CO | Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1. | TAIR | FPI, L |
| COL1 | Homologous to the flowering-time gene CONSTANS. | TAIR | FPI, L |
| COL2 | Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins | TAIR | FPI, L |
| COL3 | Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching. | TAIR | - |
| COL9 | This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO. | TAIR | L |
| COP1 | Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light. | TAIR | L |
| CRY1 | Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings. | TAIR | L |
| CRY2 | Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus. | TAIR | L |
| DPBF2 | Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons. | TAIR | - |
| E12A11 | Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed. | TAIR | - |
| EEL | Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo. | TAIR | - |
| EFS | Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development. | TAIR | V |
| ELF3 | Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions. | TAIR | L |
| ELF4 | Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes. | TAIR | L |
| ELF4-L3 | Protein of unknown function | TAIR | - |
| ELF4-L2 | Protein of unknown function | TAIR | - |
| ELF4-L3 | Protein of unknown function | TAIR | L |
| ELF4-L4 | Protein of unknown function | TAIR | - |
| ELF5 | Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC | TAIR | - |
| ELF6 | Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes. | TAIR | - |
| ELF9 | Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay. | TAIR | FPI |
| EMF1 | Involved in regulating reproductive development | TAIR | - |
| EMF2 | Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3. | TAIR | - |
| ESD4 | EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development. | TAIR | - |
| FCA | Involved in the promotion of the transition of the vegetative meristem to reproductive development. Four forms of the protein (alpha, beta, delta and gamma) are produced by alternative splicing. Involved in RNA-mediated chromatin silencing. At one point it was believed to act as an abscisic acid receptor but the paper describing that function was retracted. | TAIR | |
| FD | bZIP protein required for positive regulation of flowering. Mutants are late flowering. FD interacts with FT to promote flowering.Expressed in the shoot apex in floral anlagen, then declines in floral primordia. | TAIR | |
| FES1 | Encodes a zinc finger domain containing protein that is expressed in the shoot/root apex and vasculature, and acts with FRI to repress flowering.FES1 mutants in a Col(FRI+) background will flower early under inductive conditions. | TAIR | |
| FIE1 | Encodes a protein similar to the transcriptional regular of the animal Polycomb group and is involved in regulation of establishment of anterior-posterior polar axis in the endosperm and repression of flowering during vegetative phase. Mutation leads endosperm to develop in the absence of fertilization and flowers to form in seedlings and non-reproductive organs. Also exhibits maternal effect gametophytic lethal phenotype, which is suppressed by hypomethylation. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. In the ovule, the FIE transcript levels increase transiently just after fertilization. | TAIR | |
| FIS2 | Encodes a negative regulator of seed development in the absence of pollination. In the ovule, the FIS2 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization. | TAIR | |
| FKF1 | Encodes FKF1, a flavin-binding kelch repeat F box protein, is clock-controlled, regulates transition to flowering. Forms a complex with GI on the CO promoter to regulate CO expression. | TAIR | |
| FLC | MADS-box protein encoded by FLOWERING LOCUS C - transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock. Expression is downregulated during cold treatment. Vernalization, FRI and the autonomous pathway all influence the state of FLC chromatin. Both maternal and paternal alleles are reset by vernalization, but their earliest activation differs in timing and location. Histone H3 trimethylation at lysine 4 and histone acetylation are associated with active FLC expression, whereas histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC repression. Expression is also repressed by two small RNAs (30- and 24-nt) complementary to the FLC sense strand 3’ to the polyA site. The small RNAs are most likely derived from an antisense transcript of FLC. Interacts with SOC1 and FT chromatin in vivo. Member of a protein complex. | TAIR | |
| FLD | Encodes a plant homolog of a SWIRM domain containing protein found in histone deacetylase complexes in mammals. Lesions in FLD result in hyperacetylation of histones in FLC chromatin, up-regulation of FLC expression and extremely delayed flowering. FLD plays a key role in regulating the reproductive competence of the shoot and results in different developmental phase transitions in Arabidopsis. | TAIR | |
| FLK | RNA binding, nucleic acid binding; positive regulation of flower development | TAIR | |
| FLP1* | Encodes a small protein with unknown function and is similar to flower promoting factor 1. This gene is not expressed in apical meristem after floral induction but is expressed in roots, flowers, and in low abundance, leaves. | TAIR | |
| FPA | FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. Double mutants with FCA have reduced fertility and single/double mutants have defects in siRNA mediated chromatin silencing. | TAIR | |
| FPF1 | Encodes a small protein of 12.6 kDa that regulates flowering and is involved in gibberellin signalling pathway. It is expressed in apical meristems immediately after the photoperiodic induction of flowering. Genetic interactions with flowering time and floral organ identity genes suggest that this gene may be involved in modulating the competence to flower. There are two other genes similar to FPF1, FLP1 (At4g31380) and FLP2 (no locus name yet, on BAC F8F16 on chr 4). This is so far a plant-specific gene and is only found in long-day mustard, arabidopsis, and rice. | TAIR | |
| FRI | Encodes a major determinant of natural variation in Arabidopsis flowering time. Dominant alleles of FRI confer a vernalization requirement causing plants to overwinter vegetatively. Many early flowering accessions carry loss-of-function fri alleles .Twenty distinct haplotypes that contain non-functional FRI alleles have been identified and the distribution analyzed in over 190 accessions. The common lab strains- Col and Ler each carry loss of function mutations in FRI. | TAIR | |
| FRL1 | Encodes a sterol-C24-methyltransferases involved in sterol biosynthesis. Mutants display altered sterol composition, serrated petals and sepals and altered cotyledon vascular patterning as well as ectopic endoreduplication. This suggests that suppression of endoreduplication is important for petal morphogenesis and that normal sterol composition is required for this suppression. | TAIR | |
| FRL2 | Family member of FRI-related genes that is required for the winter-annual habit. | TAIR | |
| FT | FT, together with LFY, promotes flowering and is antagonistic with its homologous gene, TERMINAL FLOWER1 (TFL1). FT is expressed in leaves and is induced by long day treatment. Either the FT mRNA or protein is translocated to the shoot apex where it induces its own expression. Recent data suggests that FT protein acts as a long-range signal. FT is a target of CO and acts upstream of SOC1. | TAIR | |
| FUL | MADS box gene negatively regulated by APETALA1 | TAIR | |
| FVE | Controls flowering. | TAIR | |
| FY | Encodes a protein with similarity to yeast Pfs2p, an mRNA processing factor. Involved in regulation of flowering time; affects FCA mRNA processing. Homozygous mutants are late flowering, null alleles are embryo lethal. | TAIR | |
| GBF4 | Encodes a basic leucine zipper G-box binding factor that can bind to G-box motifs only as heterodimers with GBF2 or GBF3. A single amino acid change can confer G-box binding as homodimers. | TAIR | |
| GI | Together with CONSTANTS (CO) and FLOWERING LOCUS T (FT), GIGANTEA promotes flowering under long days in a circadian clock-controlled flowering pathway. GI acts earlier than CO and FT in the pathway by increasing CO and FT mRNA abundance. Located in the nucleus. Regulates several developmental processes, including photoperiod-mediated flowering, phytochrome B signaling, circadian clock, carbohydrate metabolism, and cold stress response. The gene's transcription is controlled by the circadian clock and it is post-transcriptionally regulated by light and dark. Forms a complex with FKF1 on the CO promoter to regulate CO expression. | TAIR | |
| GRF1 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower | TAIR | |
| GRF10* | Encodes a 14-3-3 protein. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1 | TAIR | |
| GRF11* | Encodes a 14-3-3 protein. Binds H+-ATPase in response to blue light. | TAIR | |
| GRF12* | Encodes a 14-3-3 protein. | TAIR | |
| GRF2 | G-box binding factor GF14 omega encoding a 14-3-3 protein | TAIR | |
| GRF3 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower. | TAIR | |
| GRF4 | GF14 protein phi chain member of 14-3-3 protein family. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1 | TAIR | |
| GRF5 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower. | TAIR | |
| GRF6 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower. | TAIR | |
| GRF7 | Encodes GF14 ν, a 14-3-3 protein isoform (14-3-3ν). | TAIR | |
| GRF8 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in shoot and flower. | TAIR | |
| HAP3A | Encodes a transcription factor from the nuclear factor Y (NF-Y) family, AtNF-YB1. Confers drought tolerance. | TAIR | |
| HAP3B | - | - | |
| HAP5A | Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5A) and expressed in vegetative and reproductive tissues. | TAIR | |
| HAP5B | Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5B) and expressed in vegetative and reproductive tissues | TAIR | |
| HAP5C | Heme activated protein (HAP5c) | TAIR | |
| HUA2 | Putative transcription factor. Member of the floral homeotic AGAMOUS pathway.Mutations in HUA enhance the phenotype of mild ag-4 allele. Single hua mutants are early flowering and have reduced levels of FLC mRNA. Other MADS box flowering time genes such as FLM and MAF2 also appear to be regulated by HUA2. HUA2 normally activates FLC expression and enhances AG function. | TAIR |