Difference between revisions of "Time of bloom"
| Line 63: | Line 63: | ||
! <i>Arabidopsis</i> Locus !! Other Names !! AA Sequence !! Pathway !! Potential Ortholog | ! <i>Arabidopsis</i> Locus !! Other Names !! AA Sequence !! Pathway !! Potential Ortholog | ||
|- | |- | ||
| − | | | + | | AT1G04400||AT-PHH1, ATCRY2, CRY2, CRYPTOCHROME 2, FHA, PHH1||[http://arabidopsis.org/servlets/TairObject?id=28374&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT1G26790||||[http://arabidopsis.org/servlets/TairObject?id=137136&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT1G29160||||[http://arabidopsis.org/servlets/TairObject?id=29883&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT2G17770||ATBZIP27, BASIC REGION/LEUCINE ZIPPER MOTIF 27, BZIP27, FD PARALOG, FDP||[http://arabidopsis.org/servlets/TairObject?id=227228&type=locus TAIR]||Am, MI|| |
|- | |- | ||
| − | | | + | | AT2G23380||CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SET1, SETDOMAIN 1, SETDOMAIN GROUP 1||[http://arabidopsis.org/servlets/TairObject?id=26534&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | | + | | AT2G32950||ARABIDOPSIS THALIANA CONSTITUTIVE PHOTOMORPHOGENIC 1, ATCOP1, CONSTITUTIVE PHOTOMORPHOGENIC 1, COP1, DEETIOLATED MUTANT 340, DET340, EMB168, EMBRYO DEFECTIVE 168, FUS1, FUSCA 1||[http://arabidopsis.org/servlets/TairObject?id=34462&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT2G34140||||[http://arabidopsis.org/servlets/TairObject?id=33865&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT2G46670*||||[http://arabidopsis.org/servlets/TairObject?id=35019&type=locus TAIR]||L|| |
| + | |- | ||
| + | | AT2G46790||APRR9, ARABIDOPSIS PSEUDO-RESPONSE REGULATOR 9, PRR9, PSEUDO-RESPONSE REGULATOR 9, TL1, TOC1-LIKE PROTEIN 1||[http://arabidopsis.org/servlets/TairObject?id=32227&type=locus TAIR]||L|| | ||
|- | |- | ||
| − | | | + | | AT2G46830||ATCCA1, CCA1, CIRCADIAN CLOCK ASSOCIATED 1||[http://arabidopsis.org/servlets/TairObject?id=32221&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT3G02380||ATCOL2, B-BOX DOMAIN PROTEIN 3, BBX3, COL2, CONSTANS-LIKE 2||[http://arabidopsis.org/servlets/TairObject?id=35635&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | | + | | AT3G07650||B-BOX DOMAIN PROTEIN 7, BBX7, COL9, CONSTANS-LIKE 9||[http://arabidopsis.org/servlets/TairObject?id=38550&type=locus TAIR]||L|| |
|- | |- | ||
| AT3G21320||||[http://arabidopsis.org/servlets/TairObject?id=39170&type=locus TAIR]||L|| | | AT3G21320||||[http://arabidopsis.org/servlets/TairObject?id=39170&type=locus TAIR]||L|| | ||
|- | |- | ||
| − | | AT3G25730||||[http://arabidopsis.org/servlets/TairObject?id=37641&type=locus TAIR]||L|| | + | | AT3G25730||EDF3, ETHYLENE RESPONSE DNA BINDING FACTOR 3||[http://arabidopsis.org/servlets/TairObject?id=37641&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT3G33520||ACTIN-RELATED PROTEIN 6, ARP6, ATARP6, EARLY IN SHORT DAYS 1, ESD1, SUF3, SUPPRESSOR OF FRI 3||[http://arabidopsis.org/servlets/TairObject?id=40458&type=locus TAIR]||Am, V|| |
|- | |- | ||
| − | | | + | | AT3G47500||CDF3, CYCLING DOF FACTOR 3||[http://arabidopsis.org/servlets/TairObject?id=36439&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT4G08920||ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, CRYPTOCHROME 1, ELONGATED HYPOCOTYL 4, HY4, OOP2, OUT OF PHASE 2||[http://arabidopsis.org/servlets/TairObject?id=129943&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/B6REW9 Grape] |
|- | |- | ||
| − | | | + | | AT4G11880||AGAMOUS-LIKE 14, AGL14||[http://arabidopsis.org/servlets/TairObject?id=129748&type=locus TAIR]||V|| |
|- | |- | ||
| − | | | + | | AT4G22950||"AGAMOUS-LIKE 19, AGL19, GL19 |
|- | |- | ||
| − | | | + | | "||[http://arabidopsis.org/servlets/TairObject?id=128336&type=locus TAIR]||V|| |
|- | |- | ||
| − | | | + | | AT4G24540||AGAMOUS-LIKE 24, AGL24||[http://arabidopsis.org/servlets/TairObject?id=127586&type=locus TAIR]||V|| |
|- | |- | ||
| − | | | + | | AT4G34530||CIB1, CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1||[http://arabidopsis.org/servlets/TairObject?id=130033&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G08230*||||[http://arabidopsis.org/servlets/TairObject?id=135669&type=locus TAIR]||V|| |
|- | |- | ||
| − | | | + | | AT5G08330||CCA1 HIKING EXPEDITION, CHE, TRANSCRIPTION FACTOR TCP21, TCP21||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||L|| |
|- | |- | ||
| − | | | + | | AT5G15840||B-BOX DOMAIN PROTEIN 1, BBX1, CO, CONSTANS, FG||[http://arabidopsis.org/servlets/TairObject?id=130492&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | | + | | AT5G15850||ATCOL1, B-BOX DOMAIN PROTEIN 2, BBX2, COL1, CONSTANS-LIKE 1||[http://arabidopsis.org/servlets/TairObject?id=130495&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | | + | | AT5G23280*||||[http://arabidopsis.org/servlets/TairObject?id=133661&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G24470||APRR5, PRR5, PSEUDO-RESPONSE REGULATOR 5||[http://arabidopsis.org/servlets/TairObject?id=135985&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G24930||ATCOL4, B-BOX DOMAIN PROTEIN 5, BBX5, COL4, CONSTANS-LIKE 4||[http://arabidopsis.org/servlets/TairObject?id=131285&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G37055||ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING||[http://arabidopsis.org/servlets/TairObject?id=500231974&type=locus TAIR]||V|| |
|- | |- | ||
| − | | | + | | AT5G39660||CDF2, CYCLING DOF FACTOR 2||[http://arabidopsis.org/servlets/TairObject?id=133414&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G48250*||B-BOX DOMAIN PROTEIN 8, BBX8||[http://arabidopsis.org/servlets/TairObject?id=133393&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G57660||ATCOL5, B-BOX DOMAIN PROTEIN 6, BBX6, COL5, CONSTANS-LIKE 5||[http://arabidopsis.org/servlets/TairObject?id=134422&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G59570*||BOA, BROTHER OF LUX ARRHYTHMO||[http://arabidopsis.org/servlets/TairObject?id=131156&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G60100||APRR3, PRR3, PSEUDO-RESPONSE REGULATOR 3||[http://arabidopsis.org/servlets/TairObject?id=133317&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G62430||CDF1, CYCLING DOF FACTOR 1||[http://arabidopsis.org/servlets/TairObject?id=131911&type=locus TAIR]||L|| |
|- | |- | ||
| − | | | + | | AT5G65050||AGAMOUS-LIKE 31, AGL31, MADS AFFECTING FLOWERING 2, MAF2||[http://arabidopsis.org/servlets/TairObject?id=135154&type=locus TAIR]||Am, FPI, V|| |
|- | |- | ||
| EFS||||[http://arabidopsis.org/servlets/TairObject?id=136429&type=locus TAIR]||V|| | | EFS||||[http://arabidopsis.org/servlets/TairObject?id=136429&type=locus TAIR]||V|| | ||
Revision as of 21:44, 5 March 2013
By Austin Mudd - Spring 2013
Contents
To Do
- Write introduction and procedure sections
- Aggregate all sequences into a single document in proper FASTA format
Introduction
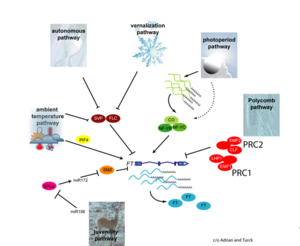
- Higgins et al., 2010
- Jung et al., 2012
- General plant anatomy / process of flowering
- Why flowering is important to study / general impact
- Factors affecting time of flowering
Procedure
- Overall procedure for finding the genes, running with GenSAS, determining SSRs, etc
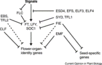
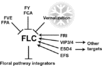
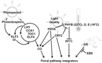
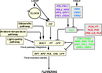
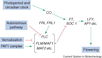
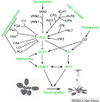
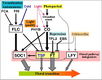
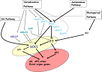
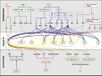
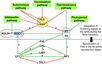
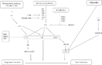
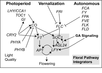
Gallery of Arabidopsis Flowering Pathways
105 Flowering Genes
The below table lists all of the genes involved in the ambient temperature, autonomous, light signaling, and vernalization pathways.
Note: All Arabidopsis genes are compiled from Jung et al., 2012. All potential orthologs are found via UniProt Grape or UniProt Strawberry nomenclature search. Formatting is the same as Jung et al., 2012 where a * indicates 1 of "24 Arabidopsis genes, which had not been previously investigated for their roles in flowering, [that] belong to [paralog groups] with known Arabidopsis flowering genes." Moreover, for pathways, "Am: Ambient temperature pathway, Au: Autonomous pathway, FPI: Flowering Pathway [Integrators], L: Light signaling pathway, MI: Meristem Identity, V: Vernalization pathway."
| Arabidopsis Locus | Other Names | AA Sequence | Pathway | Potential Ortholog |
|---|---|---|---|---|
| AT1G04400 | AT-PHH1, ATCRY2, CRY2, CRYPTOCHROME 2, FHA, PHH1 | TAIR | L | |
| AT1G26790 | TAIR | L | ||
| AT1G29160 | TAIR | L | ||
| AT2G17770 | ATBZIP27, BASIC REGION/LEUCINE ZIPPER MOTIF 27, BZIP27, FD PARALOG, FDP | TAIR | Am, MI | |
| AT2G23380 | CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SET1, SETDOMAIN 1, SETDOMAIN GROUP 1 | TAIR | Au, V | |
| AT2G32950 | ARABIDOPSIS THALIANA CONSTITUTIVE PHOTOMORPHOGENIC 1, ATCOP1, CONSTITUTIVE PHOTOMORPHOGENIC 1, COP1, DEETIOLATED MUTANT 340, DET340, EMB168, EMBRYO DEFECTIVE 168, FUS1, FUSCA 1 | TAIR | L | |
| AT2G34140 | TAIR | L | ||
| AT2G46670* | TAIR | L | ||
| AT2G46790 | APRR9, ARABIDOPSIS PSEUDO-RESPONSE REGULATOR 9, PRR9, PSEUDO-RESPONSE REGULATOR 9, TL1, TOC1-LIKE PROTEIN 1 | TAIR | L | |
| AT2G46830 | ATCCA1, CCA1, CIRCADIAN CLOCK ASSOCIATED 1 | TAIR | L | |
| AT3G02380 | ATCOL2, B-BOX DOMAIN PROTEIN 3, BBX3, COL2, CONSTANS-LIKE 2 | TAIR | FPI, L | |
| AT3G07650 | B-BOX DOMAIN PROTEIN 7, BBX7, COL9, CONSTANS-LIKE 9 | TAIR | L | |
| AT3G21320 | TAIR | L | ||
| AT3G25730 | EDF3, ETHYLENE RESPONSE DNA BINDING FACTOR 3 | TAIR | L | |
| AT3G33520 | ACTIN-RELATED PROTEIN 6, ARP6, ATARP6, EARLY IN SHORT DAYS 1, ESD1, SUF3, SUPPRESSOR OF FRI 3 | TAIR | Am, V | |
| AT3G47500 | CDF3, CYCLING DOF FACTOR 3 | TAIR | L | |
| AT4G08920 | ATCRY1, BLU1, BLUE LIGHT UNINHIBITED 1, CRY1, CRYPTOCHROME 1, ELONGATED HYPOCOTYL 4, HY4, OOP2, OUT OF PHASE 2 | TAIR | L | Grape |
| AT4G11880 | AGAMOUS-LIKE 14, AGL14 | TAIR | V | |
| AT4G22950 | "AGAMOUS-LIKE 19, AGL19, GL19 | |||
| " | TAIR | V | ||
| AT4G24540 | AGAMOUS-LIKE 24, AGL24 | TAIR | V | |
| AT4G34530 | CIB1, CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX 1 | TAIR | L | |
| AT5G08230* | TAIR | V | ||
| AT5G08330 | CCA1 HIKING EXPEDITION, CHE, TRANSCRIPTION FACTOR TCP21, TCP21 | UniProtKB | L | |
| AT5G15840 | B-BOX DOMAIN PROTEIN 1, BBX1, CO, CONSTANS, FG | TAIR | FPI, L | |
| AT5G15850 | ATCOL1, B-BOX DOMAIN PROTEIN 2, BBX2, COL1, CONSTANS-LIKE 1 | TAIR | FPI, L | |
| AT5G23280* | TAIR | L | ||
| AT5G24470 | APRR5, PRR5, PSEUDO-RESPONSE REGULATOR 5 | TAIR | L | |
| AT5G24930 | ATCOL4, B-BOX DOMAIN PROTEIN 5, BBX5, COL4, CONSTANS-LIKE 4 | TAIR | L | |
| AT5G37055 | ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING | TAIR | V | |
| AT5G39660 | CDF2, CYCLING DOF FACTOR 2 | TAIR | L | |
| AT5G48250* | B-BOX DOMAIN PROTEIN 8, BBX8 | TAIR | L | |
| AT5G57660 | ATCOL5, B-BOX DOMAIN PROTEIN 6, BBX6, COL5, CONSTANS-LIKE 5 | TAIR | L | |
| AT5G59570* | BOA, BROTHER OF LUX ARRHYTHMO | TAIR | L | |
| AT5G60100 | APRR3, PRR3, PSEUDO-RESPONSE REGULATOR 3 | TAIR | L | |
| AT5G62430 | CDF1, CYCLING DOF FACTOR 1 | TAIR | L | |
| AT5G65050 | AGAMOUS-LIKE 31, AGL31, MADS AFFECTING FLOWERING 2, MAF2 | TAIR | Am, FPI, V | |
| EFS | TAIR | V | ||
| ELF3 | TAIR | L | ||
| ELF4 | TAIR | L | ||
| ELF4-L3 | TAIR | L | ||
| FCA | TAIR | Am, Au | Grape | |
| FD | TAIR | Am, MI | ||
| FES1 | TAIR | V | ||
| FIE1 | TAIR | Au, V | ||
| FKF1 | TAIR | L | ||
| FLC | TAIR | Am, FPI, V | Grape | |
| FLD | TAIR | Am, Au | ||
| FLK | TAIR | Au | ||
| FPA | TAIR | Au | ||
| FRI | TAIR | V | ||
| FRL1 | TAIR | V | ||
| FRL2 | TAIR | V | ||
| FT | TAIR | Am, FPI | Grape, Strawberry | |
| FVE | TAIR | Am, Au | ||
| FY | TAIR | Au | ||
| GI | TAIR | L | ||
| HUA2 | TAIR | V | ||
| LD | TAIR | Au | ||
| LDL1* | TAIR | Am, Au | ||
| LDL2* | TAIR | Am, Au | ||
| LHY | TAIR | L | ||
| LKP2 | TAIR | L | ||
| LUX | TAIR | L | ||
| MAF1 | TAIR | Am, FPI, V | ||
| MAF3 | TAIR | Am, FPI, V | ||
| MAF4 | TAIR | Am, FPI, V | ||
| MAF5 | TAIR | Am, FPI, V | ||
| MEA* | TAIR | Au, V | ||
| MSI1 | TAIR | Au, V | ||
| MSI2* | TAIR | Au, V | ||
| MSI3* | TAIR | Au, V | ||
| NFC5* | TAIR | Am, Au | ||
| PAF1 | TAIR | V | ||
| PAF2 | TAIR | V | ||
| PEP | TAIR | V | ||
| PHYA | TAIR | L | Grape | |
| PHYB | TAIR | L | Grape | |
| PHYC | TAIR | L | Grape | |
| PHYD | TAIR | L | ||
| PHYE | TAIR | L | Grape | |
| PIE1 | TAIR | V | ||
| PRR7 | TAIR | L | ||
| RAV1 | TAIR | L | ||
| RFI2 | TAIR | L | ||
| SPA1 | TAIR | L | ||
| SPA2 | TAIR | L | ||
| SPA3 | TAIR | L | ||
| SPA4 | TAIR | L | ||
| SUF4 | TAIR | V | ||
| SVP | TAIR | Am, V | Grape | |
| SWN | TAIR | Au, V | ||
| TEM1 | TAIR | L | ||
| TEM2 | TAIR | L | ||
| TFL1 | TAIR | Am, FPI | Grape, Strawberry | |
| TOC1 | TAIR | L | ||
| TSF | TAIR | Am, FPI | ||
| UVR3* | TAIR | L | ||
| VEL1 | TAIR | Au, V | ||
| VEL2 | TAIR | Au | ||
| VEL3 | TAIR | Au | ||
| VIN3 | TAIR | Au, V | ||
| VRN1 | TAIR | V | ||
| VRN2 | TAIR | Au, V | ||
| VRN5 | TAIR | Au, V | ||
| ZTL | TAIR | L |