Difference between revisions of "Time of bloom"
From GcatWiki
| Line 102: | Line 102: | ||
*Procedure for setting up database | *Procedure for setting up database | ||
*Amino acid sequences for all <i>Arabidopsis</i> genes were taken from [http://www.arabidopsis.org/index.jsp The Arabidopsis Information Resource (TAIR)]. The amino acid sequences were run via tBLASTn against the blueberry scaffolds to find the closest match using the following coding: | *Amino acid sequences for all <i>Arabidopsis</i> genes were taken from [http://www.arabidopsis.org/index.jsp The Arabidopsis Information Resource (TAIR)]. The amino acid sequences were run via tBLASTn against the blueberry scaffolds to find the closest match using the following coding: | ||
| − | :<PRE>bin/tblastn -query AASequence.txt -db | + | :<PRE>bin/tblastn -query AASequence.txt -db BlueberryScaffolds.txt > AASequenceOutput.txt</PRE> |
*The best match was presumed to be the paralog of the <i>Arabidopsis</i> gene in <i>Vaccinium corymbosum</i>. A maximum E value of e-15 was established. For tBLASTn results that did fall below the E value cutoff, attempts were made to find and tBLASTn a <i>Vitis vinifera</i> paralog of the <i>Arabidopsis</i> gene from [http://www.uniprot.org/uniprot/?query=organism%3A%22Vitis+vinifera+%5B29760%5D%22&sort=score UniProtKB] nomenclature search. | *The best match was presumed to be the paralog of the <i>Arabidopsis</i> gene in <i>Vaccinium corymbosum</i>. A maximum E value of e-15 was established. For tBLASTn results that did fall below the E value cutoff, attempts were made to find and tBLASTn a <i>Vitis vinifera</i> paralog of the <i>Arabidopsis</i> gene from [http://www.uniprot.org/uniprot/?query=organism%3A%22Vitis+vinifera+%5B29760%5D%22&sort=score UniProtKB] nomenclature search. | ||
*Procedure for running GenSAS and determining SSRs | *Procedure for running GenSAS and determining SSRs | ||
Revision as of 18:09, 14 March 2013
Austin Mudd - Spring 2013
Shortened URL: http://goo.gl/zuTkP
Contents
To Do
- Continue writing the methods section
- Run amino acid sequences
- Note: <br/> works on graphs
Introduction to Flowering
The Process of Flowering
- Flowering is the "switch from vegetative growth (the production of stems and leaves) to reproductive growth (the production of flowers)" (Higgins et al., 2010)
- The “shoot apical meristem starts to produce flowers instead of leaves” (Fornara et al., 2010)
- Occurs “when conditions for pollination and seed development are optimal and consequently most plants restrict flowering to a specific time of year” (Higgins et al., 2010)
- ”The genes and molecular mechanisms controlling flowering have been extensively studied in the model dicot Arabidopsis thaliana” (Higgins et al., 2010)
- In Arabidopsis thaliana, “180 genes have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants ... Strikingly, several genes act more than once and in several tissues during floral induction” (Fornara et al., 2010)
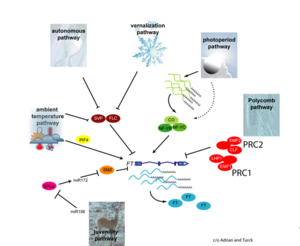
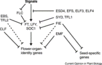
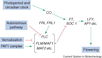
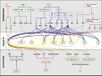
The major pathways in the timing of flowering from the Max Planck Institute for Plant Breeding Research
The Timing of Flowering
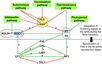
- Flowering is controlled by several “major pathways: the photoperiod and vernalization pathways control flowering in response to seasonal changes in day length and temperature; the ambient temperature pathway responds to daily growth temperatures; and the age, autonomous, and gibberellin pathways act more independently of environmental stimuli.” (Fornara et al., 2010)
- These “pathways converge to regulate a small number of ‘floral integrator genes,’ ... which govern flowering time by merging signals from multiple pathways” (Fornara et al., 2010)
The Importance of Flowering
- ”Flowering is one of the most important agronomic traits influencing crop yield” (Jung et al., 2012)
- ”Flowering time is important for adaptation to specific environments and the world's major crop species provide a particularly interesting opportunity for study because they are grown in areas outside the ecogeographical limits of their wild ancestors” (Higgins et al., 2010)
- “Adaptation to different environments and practices has been achieved by manipulation of flowering time responses” (Higgins et al., 2010)
- The study of flowering is ”critical for the breeding of climate change resilient crop varieties” (Jung et al., 2012)
- Flowering is “an excellent system for comparison between and within domestic and wild species” (Higgins et al., 2010)
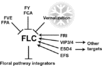
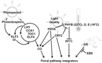
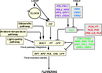
Pathways Controlling Flowering
Age Pathway
- "The miR156–SPL interaction constitutes an evolutionarily conserved, endogenous cue for both vegetative phase transition and flowering ... The age-dependent decrease in miR156 results in an increase in SPLs that promote juvenile to adult phase transition and flowering through activation of miR172, MADS box genes, and LFY" (Yu et al., 2012)
- 5 Arabidopsis genes are involved in the age pathway: SPL3, SPL4, SPL5, SPL9, SPL10 (Amasino, 2010)
Ambient Temperature Pathway
- Unlike "the photoperiod and vernalisation pathways [which] monitor seasonal changes in day length or temperature and ... [respond] to exposure to long days or prolonged cold temperatures, the ambient temperature pathway coordinates the response to daily growth temperatures" (Jung et al., 2012)
- 16 Arabidopsis genes are involved in the ambient temperature pathway: AGL31, ATARP6, ATBZIP27, FCA, FD, FLC, FLD, FT, FVE, MAF1, MAF3, MAF4, MAF5, SVP, TFL1, TSF (Jung et al., 2012)
Autonomous Pathway
- The autonomous pathway is "activated in response to endogenous changes that are independent from the environmental cues leading to flowering", such as the plant's circadian rhythm (Jung et al., 2012)
- 17 Arabidopsis genes are involved in the autonomous pathway: CLF, FCA, FIE1, FLD, FLK, FPA, FVE, FY, LD, MSI1, SWN, VEL1, VEL2, VEL3, VIN3, VRN2, VRN5 (Jung et al., 2012)
Gibberellin Pathway
- Gibberellin "is essential for floral induction in short-day conditions." In fact, plants with a "mutation in a GA biosynthetic gene, such as GA1, fail to flower" (Yu et al., 2012)
- 5 Arabidopsis genes are involved in the gibberellin pathway: GAI, GID1, RGA, RGL1, RGL2 (Yu et al., 2012)
Light Signaling Pathway
- "Light is one of the main environmental regulators of flowering in plants. Plants sense the time of day and season of year by monitoring the light environment through light signalling pathways." Furthermore, the light signalling pathway is comprised of the "photoperiod pathway genes together with photoreceptor genes and circadian clock components" (Jung et al., 2012)
- 48 Arabidopsis genes are involved in the light signaling pathway: APRR3, APRR5, APRR9, AT1G26790, AT1G29160, AT2G34140, AT3G21320, AT3G25730, ATCOL4, ATCOL5, CCA1, CDF1, CDF2, CDF3, CDF5, CHE, CIB1, CO, COL1, COL2, COL9, COP1, CRY1, CRY2, ELF3, ELF4, ELF4-L3, FKF1, GI, LHY, LKP2, LUX, PHYA, PHYB, PHYC, PHYD, PHYE, PRR7, RAV1, RFI2, SPA1, SPA2, SPA3, SPA4, TEM1, TEM2, TOC1, ZTL (Jung et al., 2012)
Polycomb Pathway
- The polycomb pathway centers on “epigenetic [repression] … [of] various developmental and cellular processes … [through two] multi-subunit protein complexes: Polycomb Repressor Complex 1 (PRC1)” and Polycomb Repressor Complex 2 (PRC2) (Kim et al., 2012)
- 10 Arabidopsis genes are involved in the polycomb pathway: CLF, EMF1, EMF2, FIE1, FIS2, LHP1, MEA, MSI1, SWN, VRN2 (Kim et al., 2012)
Vernalization Pathway
- The vernalization pathway is the response to "prolonged periods of low temperature [that are required] to initiate flowering" (Jung et al., 2012)
- 32 Arabidopsis genes are involved in the vernalization pathway: AGL14, AGL19, AGL24, AGL31, ATARP6, ATSWC6, CLF, EFS, FES1, FIE1, FLC, FRI, FRL1, FRL2, HUA2, MAF1, MAF3, MAF4, MAF5, MSI1, PAF1, PAF2, PEP, PIE1, SUF4, SVP, SWN, VEL1, VIN3, VRN1, VRN2, VRN5 (Jung et al., 2012)
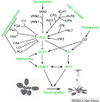
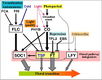
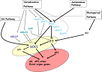
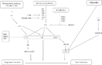
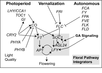
Gallery of Arabidopsis Flowering Pathways
Methods
Finding Genes
- I examined a variety of journal articles related to time of flowering in Arabidopsis thaliana and found a number of pathways related to flowering (see the gallery above). I came across a genomic analysis of soybean by Jung et al., 2012. In this paper, they listed the "183 Arabidopsis genes that are known to take part in flowering regulatory pathways [taken] from previous studies." These 183 genes, plus "24 additional Arabidopsis genes that are grouped into the same [homolog groups] as known flowering genes," provided a solid foundation for my study. (Jung et al., 2012)
- These 207 total genes fell into two categories: 1) flowering pathway integrators/meristem identity genes and 2) condition pathway genes (responding to the photoperiod pathway, the vernalization pathway, the ambient temperature pathway, the autonomous pathway, and other pathways). Per the direction of Dr. Jeannie Rowland of the USDA Genetic Improvement for Fruits and Vegetables Laboratory, I focused on the condition pathway genes.
- I identified a total of seven different pathways controlling flowering: the age pathway, the ambient temperature pathway, the autonomous pathway, the gibberellin pathway, the light signaling pathway, the polycomb pathway, and the vernalization pathway. Descriptions and the primary genes involved in these pathways were taken from Amasino, 2010, Jung et al., 2012, Kim et al., 2012, and Yu et al., 2012.
- A total of 61 genes were examined, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." (Fornara et al., 2010) Due to limited time, however, the genes involved in the light signaling pathway were not examined.
Finding SSRs
- Procedure for setting up database
- Amino acid sequences for all Arabidopsis genes were taken from The Arabidopsis Information Resource (TAIR). The amino acid sequences were run via tBLASTn against the blueberry scaffolds to find the closest match using the following coding:
bin/tblastn -query AASequence.txt -db BlueberryScaffolds.txt > AASequenceOutput.txt
- The best match was presumed to be the paralog of the Arabidopsis gene in Vaccinium corymbosum. A maximum E value of e-15 was established. For tBLASTn results that did fall below the E value cutoff, attempts were made to find and tBLASTn a Vitis vinifera paralog of the Arabidopsis gene from UniProtKB nomenclature search.
- Procedure for running GenSAS and determining SSRs
Previous Examined Genes
Flowering Genes Results
This table lists 61 genes involved in the age, ambient temperature, autonomous, gibberellin, light signaling, polycomb, and vernalization pathways, almost all of which "have been implicated in flowering-time control based on isolation of loss-of-function mutations or analysis of transgenic plants." (Fornara et al., 2010)
| Arabidopsis Locus | Other Names | AA Source | Pathway |
|---|---|---|---|
| AT1G02580 | EMB173, EMBRYO DEFECTIVE 173, FERTILIZATION INDEPENDENT SEED 1, FIS1, MEA, MEDEA, SDG5, SET DOMAIN-CONTAINING PROTEIN 5 | TAIR | Polycomb |
| AT1G14920 | GAI, GIBBERELLIC ACID INSENSITIVE, RESTORATION ON GROWTH ON AMMONIA 2, RGA2 | TAIR | Gibberellin |
| AT1G20330 | COTYLEDON VASCULAR PATTERN 1, CVP1, FRILL1, FRL1, SMT2, STEROL METHYLTRANSFERASE 2 | TAIR | Vernalization |
| AT1G27370 | SPL10, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 10 | TAIR | Age |
| AT1G30970 | SUF4, SUPPRESSOR OF FRIGIDA4 | TAIR | Vernalization |
| AT1G31814 | FRIGIDA LIKE 2, FRL2 | TAIR | Vernalization |
| AT1G47250 | 20S PROTEASOME ALPHA SUBUNIT F2, PAF2 | TAIR | Vernalization |
| AT1G53160 | FLORAL TRANSITION AT THE MERISTEM6, FTM6, SPL4, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4 | TAIR | Age |
| AT1G65480 | FLOWERING LOCUS T, FT | TAIR | Ambient Temperature |
| AT1G66350 | RGA-LIKE 1, RGL, RGL1 | TAIR | Gibberellin |
| AT1G77080 | AGAMOUS-LIKE 27, AGL27, FLM, FLOWERING LOCUS M, MADS AFFECTING FLOWERING 1, MAF1 | TAIR | Ambient Temperature, Vernalization |
| AT1G77300 | ASH1 HOMOLOG 2, ASHH2, CAROTENOID CHLOROPLAST REGULATORY1, CCR1, EARLY FLOWERING IN SHORT DAYS, EFS, LAZ2, LAZARUS 2, SDG8, SET DOMAIN GROUP 8 | TAIR | Vernalization |
| AT2G01570 | REPRESSOR OF GA, REPRESSOR OF GA1-3 1, RGA, RGA1 | TAIR | Gibberellin |
| AT2G17770 | ATBZIP27, BASIC REGION/LEUCINE ZIPPER MOTIF 27, BZIP27, FD PARALOG, FDP | TAIR | Ambient Temperature |
| AT2G18870 | VEL3, VERNALIZATION5/VIN3-LIKE 3, VIL4, VIN3-LIKE 4 | TAIR | Autonomous |
| AT2G18880 | VEL2, VERNALIZATION5/VIN3-LIKE 2, VIL3, VIN3-LIKE 3 | TAIR | Autonomous |
| AT2G19520 | ACG1, ATMSI4, FVE, MSI4, MULTICOPY SUPPRESSOR OF IRA1 4, NFC04, NFC4 | TAIR | Ambient Temperature, Autonomous |
| AT2G22540 | AGAMOUS-LIKE 22, AGL22, SHORT VEGETATIVE PHASE, SVP | TAIR | Ambient Temperature, Vernalization |
| AT2G23380 | CLF, CURLY LEAF, ICU1, INCURVATA 1, SDG1, SET1, SETDOMAIN 1, SETDOMAIN GROUP 1 | TAIR | Autonomous, Polycomb, Vernalization |
| AT2G33810 | SPL3, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 | TAIR | Age |
| AT2G33835 | FES1, FRIGIDA-ESSENTIAL 1 | TAIR | Vernalization |
| AT2G35670 | FERTILIZATION INDEPENDENT SEED 2, FERTILIZATION-INDEPENDENT ENDOSPERM 2, FIE2, FIS2 | TAIR | Polycomb |
| AT2G42200 | ATSPL9, SPL9, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 | TAIR | Age |
| AT2G43410 | FPA | TAIR | Autonomous |
| AT3G03450 | RGA-LIKE 2, RGL2 | TAIR | Gibberellin |
| AT3G04610 | FLK, FLOWERING LOCUS KH DOMAIN | TAIR | Autonomous |
| AT3G05120 | ATGID1A, GA INSENSITIVE DWARF1A, GID1A | TAIR | Gibberellin |
| AT3G10390 | FLD, FLOWERING LOCUS D | TAIR | Ambient Temperature, Autonomous |
| AT3G12810 | CHR13, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1, PIE1, SRCAP | TAIR | Vernalization |
| AT3G15270 | SPL5, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5 | TAIR | Age |
| AT3G18990 | REDUCED VERNALIZATION RESPONSE 1, REM39, REPRODUCTIVE MERISTEM 39, VRN1 | TAIR | Vernalization |
| AT3G20740 | FERTILIZATION-INDEPENDENT ENDOSPERM, FERTILIZATION-INDEPENDENT ENDOSPERM 1, FIE, FIE1, FIS3 | TAIR | Autonomous, Polycomb, Vernalization |
| AT3G24440 | VERNALIZATION 5, VIL1, VIN3-LIKE 1, VRN5 | TAIR | Autonomous, Vernalization |
| AT3G33520 | ACTIN-RELATED PROTEIN 6, ARP6, ATARP6, EARLY IN SHORT DAYS 1, ESD1, SUF3, SUPPRESSOR OF FRI 3 | TAIR | Ambient Temperature, Vernalization |
| AT4G00650 | FLA, FLOWERING LOCUS A, FRI, FRIGIDA | TAIR | Vernalization |
| AT4G02020 | EZA1, SDG10, SET DOMAIN-CONTAINING PROTEIN 10, SWINGER, SWN | TAIR | Autonomous, Polycomb, Vernalization |
| AT4G02560 | LD, LUMINIDEPENDENS | TAIR | Autonomous |
| AT4G11880 | AGAMOUS-LIKE 14, AGL14 | TAIR | Vernalization |
| AT4G16280 | FCA | TAIR | Ambient Temperature, Autonomous |
| AT4G16845 | REDUCED VERNALIZATION RESPONSE 2, VRN2 | TAIR | Autonomous, Polycomb, Vernalization |
| AT4G20370 | TSF, TWIN SISTER OF FT | TAIR | Ambient Temperature |
| AT4G22950 | AGAMOUS-LIKE 19, AGL19, GL19 | TAIR | Vernalization |
| AT4G24540 | AGAMOUS-LIKE 24, AGL24 | TAIR | Vernalization |
| AT4G26000 | PEP, PEPPER | TAIR | Vernalization |
| AT4G30200 | VEL1, VERNALIZATION5/VIN3-LIKE 1, VIL2, VIN3-LIKE 2 | TAIR | Autonomous, Vernalization |
| AT4G35900 | ATBZIP14, FD, FD-1 | TAIR | Ambient Temperature |
| AT5G03840 | TERMINAL FLOWER 1, TFL1 | TAIR | Ambient Temperature |
| AT5G10140 | AGAMOUS-LIKE 25, AGL25, FLC, FLF, FLOWERING LOCUS C, FLOWERING LOCUS F | TAIR | Ambient Temperature, Vernalization |
| AT5G11530 | EMBRYONIC FLOWER 1, EMF1 | TAIR | Polycomb |
| AT5G13480 | FY | TAIR | Autonomous |
| AT5G17690 | ATLHP1, LHP1, LIKE HETEROCHROMATIN PROTEIN 1, TERMINAL FLOWER 2, TFL2 | TAIR | Polycomb |
| AT5G23150 | ENHANCER OF AG-4 2, HUA2 | TAIR | Vernalization |
| AT5G37055 | ATSWC6, SEF, SERRATED LEAVES AND EARLY FLOWERING | TAIR | Vernalization |
| AT5G42790 | ARS5, ARSENIC TOLERANCE 5, ATPSM30, PAF1, PROTEASOME ALPHA SUBUNIT F1 | TAIR | Vernalization |
| AT5G51230 | ATEMF2, CYR1, CYTOKININ RESISTANT 1, EMBRYONIC FLOWER 2, EMF2, VEF2 | TAIR | Polycomb |
| AT5G57380 | VERNALIZATION INSENSITIVE 3, VIN3 | TAIR | Autonomous, Vernalization |
| AT5G58230 | ARABIDOPSIS MULTICOPY SUPRESSOR OF IRA1, ATMSI1, MATERNAL EFFECT EMBRYO ARREST 70, MEE70, MSI1, MULTICOPY SUPRESSOR OF IRA1 | TAIR | Autonomous, Polycomb, Vernalization |
| AT5G65050 | AGAMOUS-LIKE 31, AGL31, MADS AFFECTING FLOWERING 2, MAF2 | TAIR | Ambient Temperature, Vernalization |
| AT5G65060 | AGAMOUS-LIKE 70, AGL70, FCL3, MADS AFFECTING FLOWERING 3, MAF3 | TAIR | Ambient Temperature, Vernalization |
| AT5G65070 | AGAMOUS-LIKE 69, AGL69, FCL4, MADS AFFECTING FLOWERING 4, MAF4 | TAIR | Ambient Temperature, Vernalization |
| AT5G65080 | AGAMOUS-LIKE 68, AGL68, MADS AFFECTING FLOWERING 5, MAF5 | TAIR | Ambient Temperature, Vernalization |