Difference between revisions of "Karen's Assignment"
MaCampbell (talk | contribs) |
MaCampbell (talk | contribs) |
||
| Line 1: | Line 1: | ||
| − | '''Potassium homeostasis''' (Download PPT Presentation) | + | '''Potassium homeostasis''' ([http://www.example.com Download PPT Presentation) |
'''How does our species maintain potassium homeostasis?''' | '''How does our species maintain potassium homeostasis?''' | ||
Revision as of 13:19, 1 October 2009
Potassium homeostasis ([http://www.example.com Download PPT Presentation)
How does our species maintain potassium homeostasis?
In JGI's list of genes with predicted functions, I found the following 10 genes related to K+ (6 of which were called in RAST):
K+ transport system, NAD-binding component
2603205..2603861(+)
Trk system potassium uptake protein trkA-1
Start 2603205
Stop 2603861
This is the same as a JGI called gene.
K+ transport system, NAD-binding component
2199065..2199751(+)
Trk system potassium uptake protein trkA-3
Start 2199116
Stop 2199751
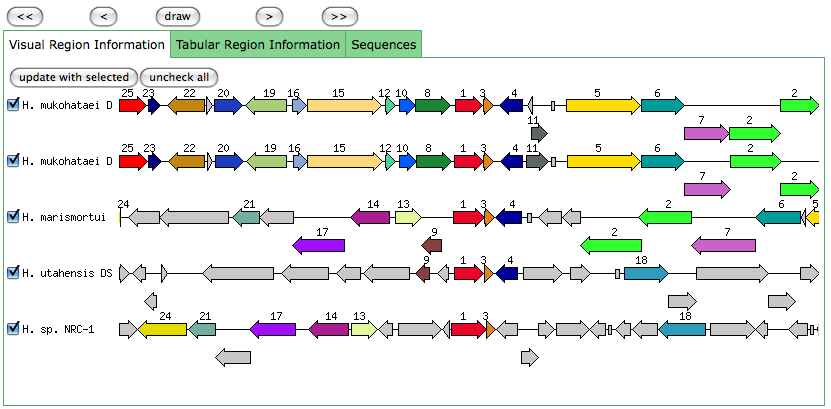
Similar to a JGI called gene, but the start position is later in RAST than JGI.
K+ transport system, NAD-binding component
2210390..2211088(-)
Kef-type K+ ransport system, predicted NAD-binding component
2271340..2272527(+)
Kef-type K+ transport system, membrane component
2954301..2955512(+)
K+ transport system, NAD-binding component
1938027..1939364(-)
Trk system potassium uptake protein trkA-4
Start 1939364
Stop 1938027
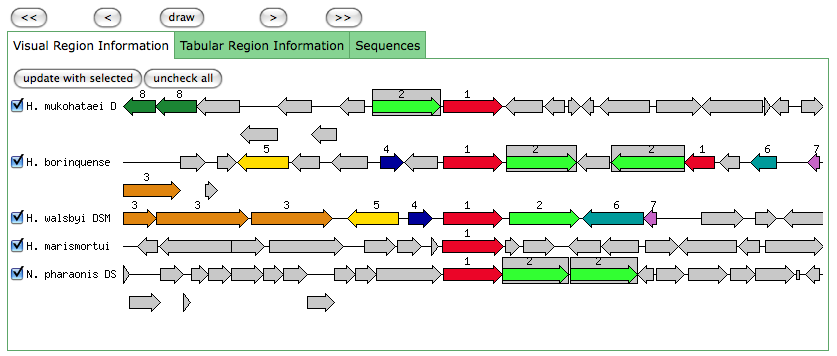
This is the same as a JGI called gene.
Trk-type K+ transport system, membrane component
1939429..1940991(-)
Start 1940991
Stop 1939429
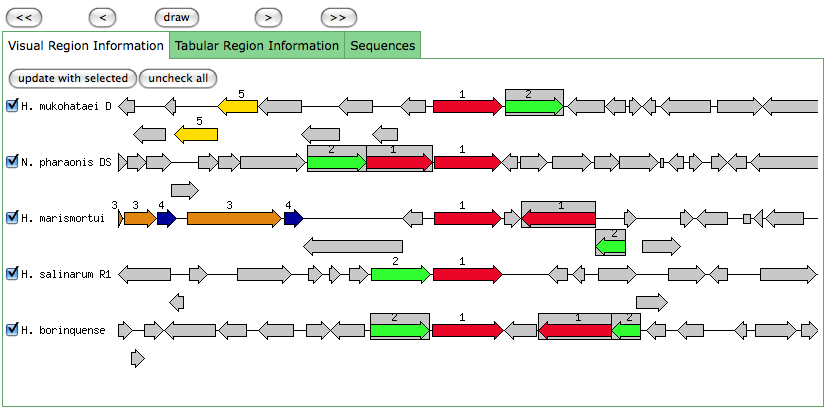
This is the same as a JGI called gene.
K+ transport system, NAD-binding component
2797145..2798779(-)
Start 2798779
Stop 2797145
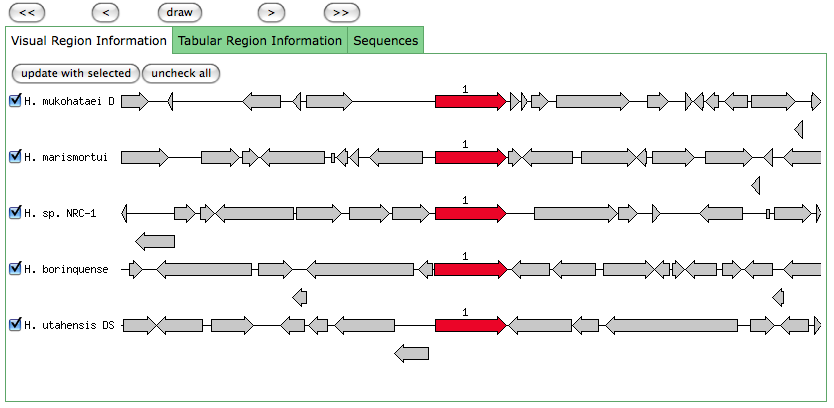
This is the same as a JGI called gene.
K+ transport system, NAD-binding component
1146592..1148280(-)
NhaP-type Na+(K+)/H+ antiporter
3021102..3022994(+)
Trk system potassium uptake protein trkA-2
Start 3021102
Stop 3022994
This is the same as a JGI called gene.
I found no genes with predicted functions that are associated with Cl- from JGI
In RAST, I found seven genes related to potassium homeostasis that were not called in JGI:
Cobalt-zinc-cadmium resistance protein
Hydroxyacylglutathione hydrolase (EC 3.1.2.6)-1
Hydroxyacylglutathione hydrolase (EC 3.1.2.6)-2
I found no genes with predicted functions that are associated with Cl- function on RAST.
Are the genes for this process highly conserved among halophiles?
Blast results for JGI genes:
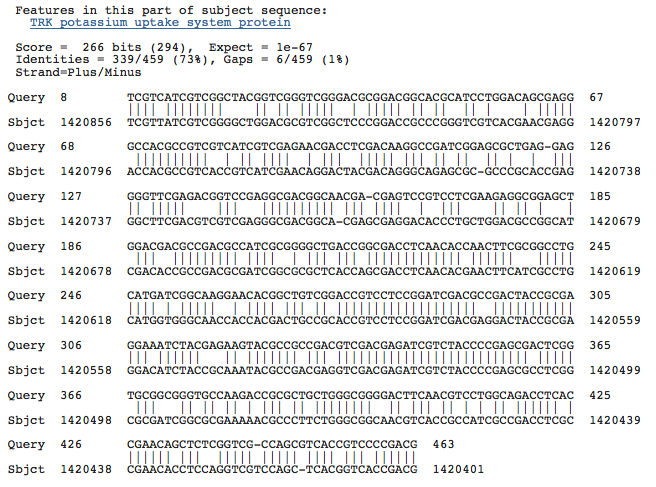
Blastn results for K+ transport system, NAD-binding component
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence

Halobacterium salinarum complete genome, strain R1
Halobacterium sp. NRC-1, complete genome
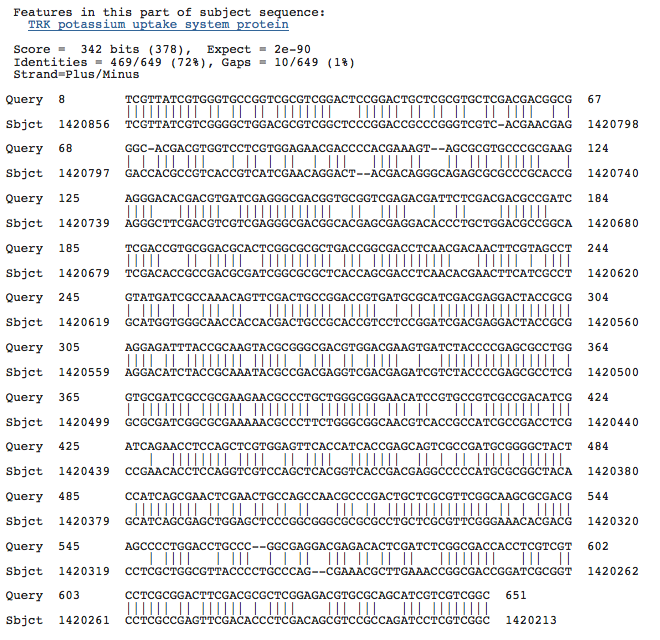
Blastn results for K+ transport system, NAD-binding component
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence
Halobacterium salinarum complete genome, strain R1
Halobacterium sp. NRC-1, complete genome
Blastn results for K+ transport system, NAD-binding component
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence
Blastn results for Kef-type K+ ransport system, predicted NAD-binding component
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence
Halorubrum lacusprofundi ATCC 49239 chromosome 1, complete sequence

Halobacterium salinarum complete genome, strain R1
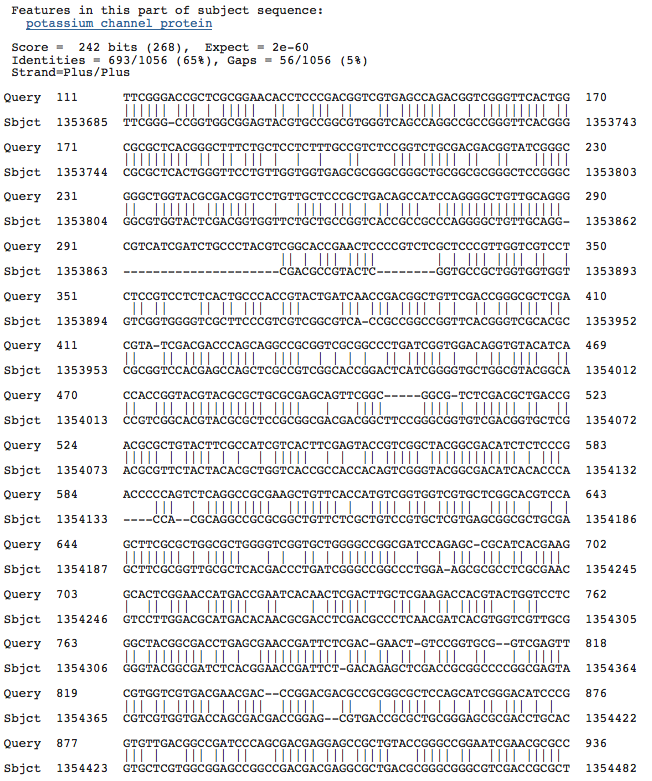
Halobacterium sp. NRC-1, complete genome

Natronomonas pharaonis DSM 2160 complete genome

Blastn results for Kef-type K+ transport system, membrane component
Halorubrum lacusprofundi ATCC 49239 chromosome 1, complete sequence
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence
Blastn results for K+ transport system, NAD-binding component
Halorubrum lacusprofundi ATCC 49239 chromosome 1, complete sequence
Natronomonas pharaonis DSM 2160 complete genome

Blastn results for Trk-type K+ transport system, membrane component
Natronomonas pharaonis DSM 2160 complete genome
[Halorubrum lacusprofundi ATCC 49239 chromosome 1, complete sequence http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=nucleotide&dopt=GenBank&RID=BJ9VZPHM01N&log%24=nuclalign&blast_rank=3&list_uids=222451341]

Haloarcula marismortui ATCC 43049 chromosome I, complete sequence

Halobacterium salinarum PHS3 plasmid complete genome, strain


Blastn results for K+ transport system, NAD-binding component
Haloarcula marismortui ATCC 43049 chromosome I, complete sequence
Halobacterium salinarum complete genome, strain R1

Halobacterium sp. NRC-1, complete genome

Halorubrum lacusprofundi ATCC 49239 chromosome 1, complete sequence


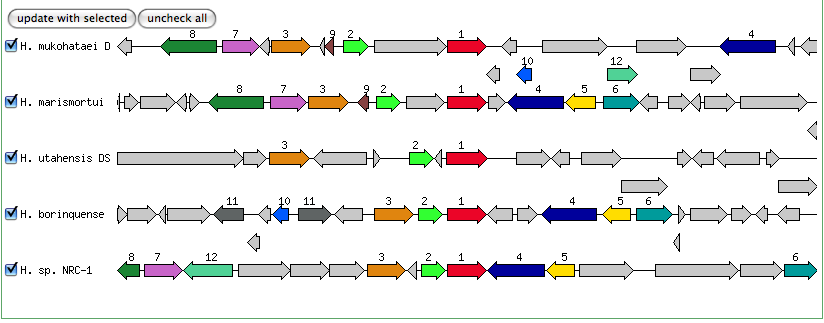
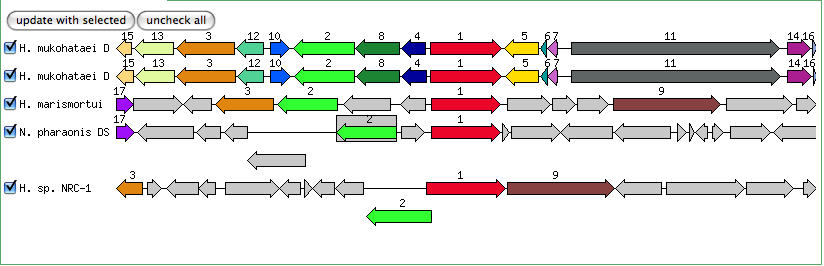
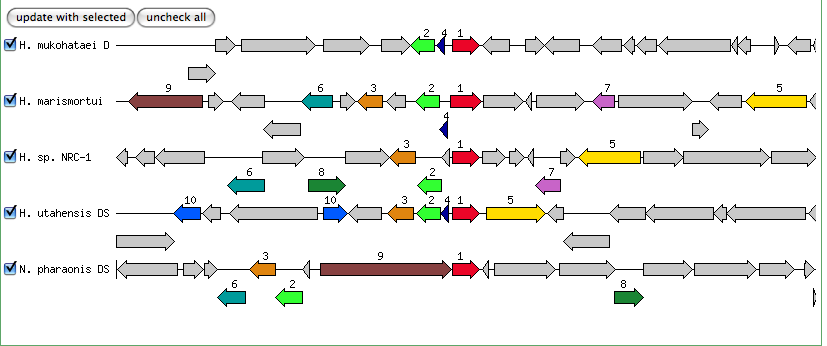
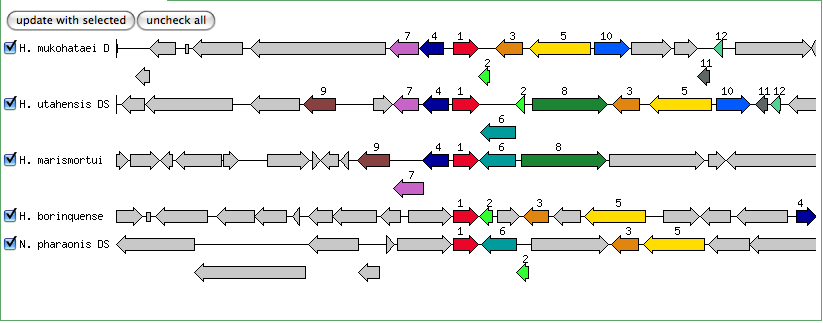
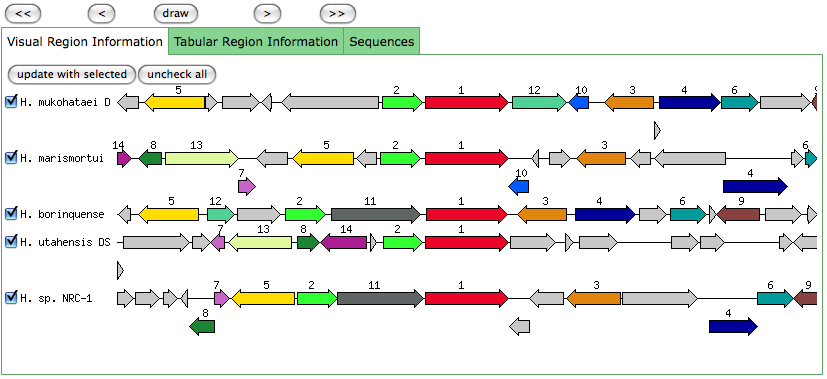
The blastn results are becoming very repetitive. I think at this point, the way to attack this problem may be to look into this list of 5-6 species that seem to share the potassium proteins seen in our species. If I can find a common thread in how other species are utilizing the same potassium homeostasis machinery, it will tell me a great deal about how our own species is using all these potassium related genes.
It is important to note that many of these genes were also related to the species Halorhabdus utahensis. I did not include the blastn results for this species because Blast could not specify which gene segment these matches were a part of. I believe this was not included simply because this species has not been annotated (or the annotation has not been entered into blast). In any comparison of species that takes place after this, I will need to include this species as well.
Is potassium homeostasis related to our species ability to survive/thrive in high salinity environments? How?
"Cell membranes are freely permeable to water, so the only way to prevent the loss of cellular water under high salt conditions is to increase the internal solute concentration. Halotolerantand halophilic microorganisms therefore accumulate high solute concentrations within the cytoplasm." [1]
I am beginning to understand potassium homeostasis and how it may be useful to our organism to survive in high salt concentrations by first researching how other known species utilize this mechanism. In a paper by Strahl and Greie, the Halobacterium salinarum species use of potassium homeostasis to maintain life in a high salinity environment is described. Apparently these organisms maintain life in such conditions by creating an equimolar condition within their cytoplasm. KCl is the preferred ion to create this equal osmolality for this halobacterium resulting in osmotic equilibrium with the surrounding environment. The K+ ion enters the cell by both passive transport mechanisms as well as active transport mechanisms. A gene, kdpFABC, has been discovered in this organism that codes for homologs of a bacterial ATP-driven K+ uptake system. Deletion of this gene is detrimental to organism survival in limiting K+ conditions. This paper also mentions that this mechanism is unique to this and another species of halobacterium because other halophiles utilize organic solutes to equalize osmotic pressures in high salinity environments. I must determine whether or not potassium homeostasis could be a mechanism of maintaining osmotic equilibrium in our species.
A paper by Oren et al. describes another species, Salinibacter ruber, that appears to be capable of surviving in high salinity environments due to the uptake of high concentrations of potassium. The finding that potassium homeostasis seems to be the mechanism of maintaining life in a high salinity environment was determined by measuring K+, Cl+, glutamate, glycine betaine, and N-alpha-acetyllysine concentrations within the cytoplasm. High amounts of K+ and Cl+ were found within the cytoplasm but low amounts of glutamate, glycine betaine, and N-alpha-acetyllysine were discovered. These three organic solutes are often associated with maintaining osmotic equilibrium in archaic halophiles that exist in high salinity environments. It is unusual that this species does not utilize these organic solutes to maintain homeostasis but instead seems to be utilizing the inorganic molecule KCl. This paper provides another example of potassium concentrations being utilized to maintain life in high salinity environments. I need to determine if our species is using this inorganic molecule or the more common organic molecules to maintain osmotic balance in a high salinity environment.
H. salinarum K+ transport mechanism comparison Found the nucleotide sequence for the KdpFABC operon in the species H. salinarum using NCBI. Compared this sequence to our species genome using Blastx. Found three significant hits:
heavy metal translocating P-type ATPase-1
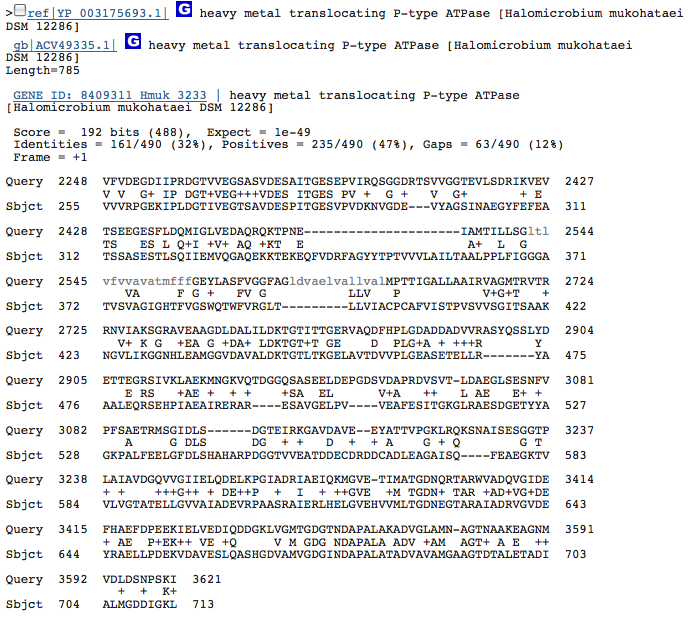
heavy metal translocating P-type ATPase-2
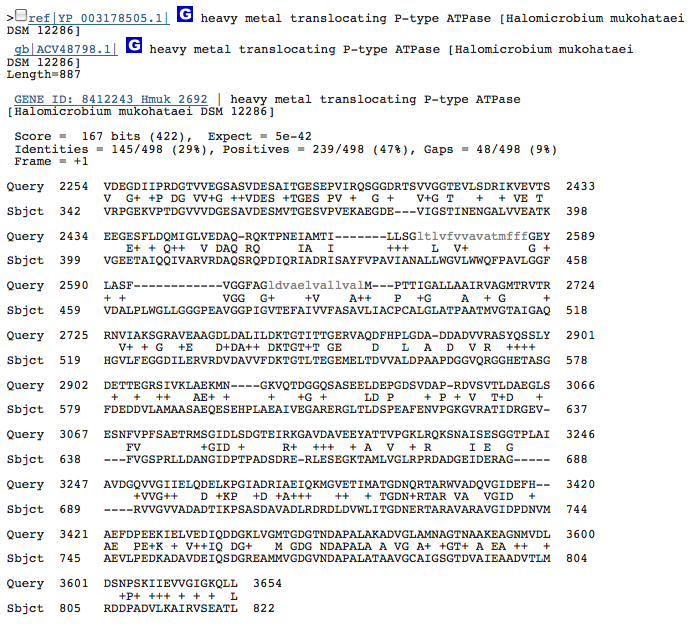
heavy metal translocating P-type ATPase-3
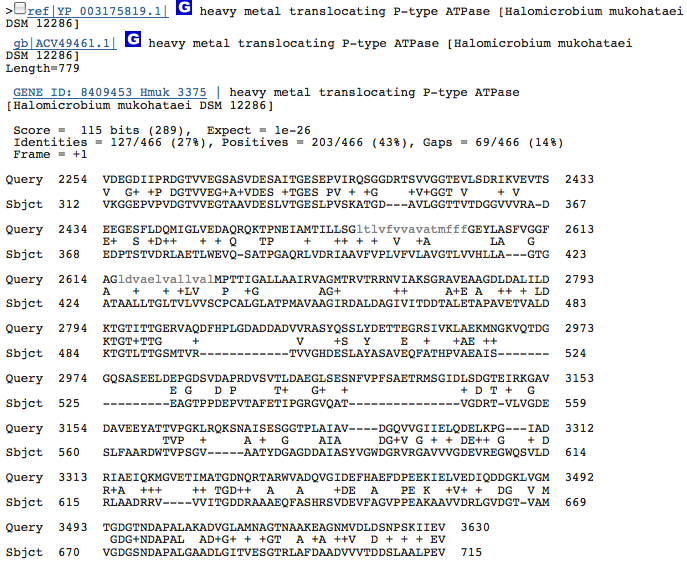
These hits could just be related to the ATPase function of the Kdp operon in H. salinarum and have nothing to do with potassium transport. I am going to redo this blastx with individual genes from operon Kdp compared to our species genome separately. When blasted separately, I found that the three hits from above were in the gene KdpB in the operon of H salinarum. No other similarities, however, were discovered.
Other species potassium uptake systems For the 6 other species that have similar potassium related genes to our species (Halorhabdus utahensis, Halorubrum lacusprofundi, Halobacterium sp. NRC-1, Haloarcula marismortui, Natronomonas pharaonis, & Halogeometricum borinquense. Excluding Halobacterium salinarum since I already compared the KdpFABC operon), I reviewed literature and did pubmed searches trying to find anything that would relate potassium to these species. The only time I found potassium mentioned in conjunction with one of these species was when one website said that Halobacterium sp. NRC-1 has potassium uptake systems coded for in its genome Kenyon MicrobeWiki. Since this was the only time potassium was mentioned in conjunction with any of these species, it is unlikely that potassium is essential for the survival of these species. Because of this fact and the lack of evidence of the KdpFABC operon in our species genome, it seems unlikely that potassium homeostasis is essential for our species survival in high salinity environments.