Difference between revisions of "Davidson Missouri W/Gene splitting"
| Line 113: | Line 113: | ||
-We did not consider any Amino Acids near the N or C terminus | -We did not consider any Amino Acids near the N or C terminus | ||
| − | -We did not consider any residues near ß-sheets or ∂-helices close to the active site because hydrogen bonding plays an active role in substrate stabilization and the polarity of our hix site could disrupt the secondary structure and therefore the hydrogen bonding ability of KNTase) || graphic | + | -We did not consider any residues near ß-sheets or ∂-helices close to the active site because hydrogen bonding plays an active role in substrate stabilization and the polarity of our hix site could disrupt the secondary structure and therefore the hydrogen bonding ability of KNTase) |
| + | | | ||
| + | |align="center"| graphic | ||
|- | |- | ||
Revision as of 15:49, 28 June 2007
We have selected 4 genes to split. We will use our online gene splitting tool to choose the PCR primers. Davidson will produce these 4 split genes and test each one.
- Red Fluorescent Protein
- Kanamycin Nucleotidyltransferase
- Cre Recombinase
- Chloramphenicol Acetyltransferase
We will also produce two constructs for tetsing promoters. MWSU will produec (Kan, RFP, Tet) while Davidson will produce (Kan, Tet, RFP). We can drop in different promoters and look for phenotypes.
Davidson is also going to have synthesized an improved pLac promoter that is shorter, will have better repression, better induction, and lack the backwards promotion we have detected with Part: BBa_R0010. We will test out the modified promoter BBa_R0011 which is reported to have good repression and strong induction. We may still introduce the UV5 double mutation to enhance transcription and compare with R0010.
MWSU is also going to produce backwards LacIq to put upstream of pLac BBa_R0010. The purpose of this is to have more LacIq in the cytoplasm at all times, regardless of ITPG status.
MWSU is also testing two of the one-time-flippable HixC sites produced by Bruce spring 2007.
MWSU is going to produce a new Hin-LVA expression plasmid that has ColE1 ori and uses Tet as the selectable marker.
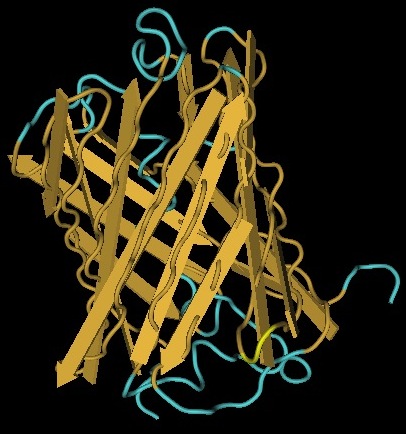
Kanamycin Nucleotidyltransferase
The yellow bands at the top and bottom of the molecule denotes hix site insertion
We decided to insert our hix sites at the 125 AA of each monomer due to their distance from each other, active site secondary structure, N or C terminus, and lack of any previous mutational analysis proving its function as integral.
Chloramphenicol Acetyltransferase
Chloramphenicol Acetyltransferase was one of the genes we chose as a node for our Hamiltonian Path. It is a bacterial gene that neutralizes the effect of an antibiotic, Chloramphenicol, by transferring acetyl groups to Chloramphenicol and changes its shape into a harmless form.
The specific Chloramphenicol Acetyltransferase gene we are using comes from the plasmid PSV2CAT whose original source is an E. coli transposable element Tn9 (Sambrook, 2001) Its PDB ID# is 1PD5.
I have chosen to insert my hixC site between amino acid 52 and 53. I chose this point because it is away from the active site of the protein, the point that contains the catalytic binding sites and allow the recognition and binding of the substrate. It is important for the insertion point to be away from the active site because we do not want the overall function and structure of the protein to be destroyed in the process of splitting. We want to split at a point where the two halves of the protein cannot work as single units, but once a hixC site has been inserted, and the two halves are brought back together, the protein displays its original function.
The structure of a type I Chloramphenicol Acetyltransferase used in the BioBrick Registry.
The structure of a type III Chloramphenicol Acetyltransferase.
This structure shows the active sites of the protein. Although the two proteins are of different types and the amino acid sequence are different, the structures are similar and we can therefore see that the point where we have decided to split is “safe”. The exact points that I have decided to cut is away from the active site.
Cre Recombinase
File:Cre recombinase monomer.png
File:Cre recombinase tetramer.png
[1].
| Gene | Description | Graphic | |||
|---|---|---|---|---|---|
| Kanamycin Nucleotidyltransferase | One gene our team will be using as a node in our Hamiltonian Path problem is Kanamycin resistance translated in the form of Kanamycin nucleotidyltransferase (KNTase). The antibiotic Kanamycin, once in the cytosol of E.Coli, inhibits protein synthesis by interacting with the “decoding” region in the small ribosomal subunit RNA.(Sambrook and Russel, 2001) The KNTase enzyme, as a member of the aminoglycoside phosphotransferase (APH) enzyme family, blocks Kanamycin’s ability to inhibit protein synthesis by transferring a nucleoside monophosphate (adenyl) group from Mg2+-ATP to the 4’ hydroxyl group of Kanamycin, inhibiting its ability to bind to the srRNA.1
Our goal was to insert a hix site (a polar molecule) in an area of KNTase protein that would not interfere with its ability to inhibit Kanamycin. We looked at mutational analysis of KNTase and other aminoglycoside phosphotransferase enzymes to determine which aspects of KNTase’s structure were integral to its function and therefore not an ideal site for hix site insertion. KNTase is a dimmer consisting of 253 amino acids in the molecule 3. In looking at conserved structures in the APH family we took into consideration that: -Substitution of AA 190 caused 650-fold decrease in enzyme activity 2 -AA 190 is involved in catalysis 5 -AA 195 and 208 are involved in Mg2+ binding 5 -Mutant Enzymes 190, 205, 210 all showed changes in mg+2 binding from the WT 2 -Substitution of AA 210 (conserved) reduced enzyme activity 2 -AA 166 serves to catalyze reactions involving ATP 2 -AA 44 is involved in ATP binding 5 -AA 60 is involved in orientation of AA 44 and ATP binding 5 -We did not consider any Amino Acids near the N or C terminus -We did not consider any residues near ß-sheets or ∂-helices close to the active site because hydrogen bonding plays an active role in substrate stabilization and the polarity of our hix site could disrupt the secondary structure and therefore the hydrogen bonding ability of KNTase) |
graphic | |||
| DsRed - Red Fluorescent Protein |
We use genes to represent the nodes on our Hamiltonian path. One of the essential features of these genes is that they can tolerate the insertion of a Hix site. It has been previously demonstrated that GFP fluoresces despite a Hix insertion. Another glowing protein, RFP (from Discosoma sp.), is a candidate for use in our path. Although its DNA sequence is markedly different from GFP's, it has some amino acid similarity and a remarkable structural similarity. Both proteins have a Beta-barrel structure which surrounds an internal chromophore. Inserting 13 amino acids into a protein can potentially disrupt its ability to function. It is thus essential to find an insertion point that does not interfere with the protein's function. Fortunately, the similarity between GFP and RFP allows us to make a highly educated guess for where to insert. RFP's amino acid position 154 is homologous to GFP's amino acid position 157, which is where GFP was split. This is therefore our best guess for where to insert the Hix site. | ||||