Difference between revisions of "Time of bloom"
(→Arabidopsis Flowering Genes) |
|||
| Line 28: | Line 28: | ||
===<i>Arabidopsis</i> Flowering Genes=== | ===<i>Arabidopsis</i> Flowering Genes=== | ||
| − | Note: All genes are compiled from [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung et al., 2012]. Formatting is the same as | + | Note: All genes are compiled from [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung et al., 2012]. Formatting is the same as [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0038250 Jung et al., 2012] where a * indicates 1 of "24 <i>Arabidopsis</i> genes, which had not been previously investigated for their roles in flowering, [that] belong to [ortholog groups] with known <i>Arabidopsis</i> flowering genes." Moreover, for pathways, "Am: Ambient temperature pathway, Au: Autonomous pathway, FPI: Flowering Pathway Intergrators, L: Light signaling pathway, MI: Meristem Identity, V: Vernalization pathway." |
{| class="wikitable" | {| class="wikitable" | ||
Revision as of 19:44, 14 February 2013
Contents
To Do
Note all Arabidopsis genes in File:Jung 207 Arabidopsis Flowering Genes.pdf from Jung et al., 2012.
Look for strawberry orthologs of Arabidopsis genes, such as in Mouhu et al., 2009.
Arabidopsis Flowering Pathway (2004-Present)
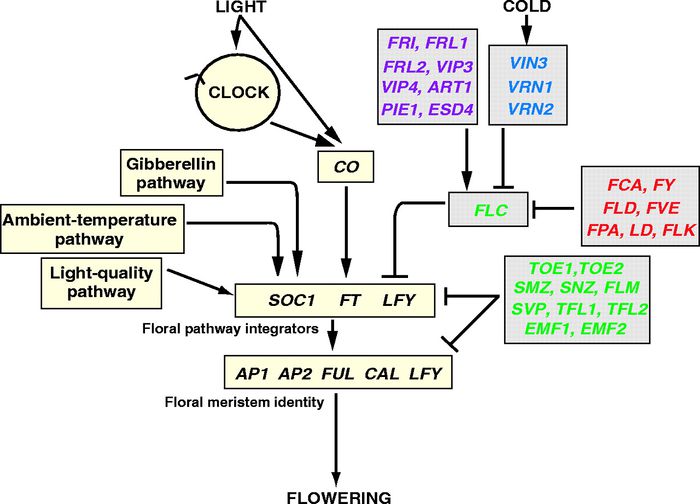
Overall pathway for the time of bloom from Henderson and Dean, 2004.
Overall pathway for the time of bloom from Schneitz, 2009.
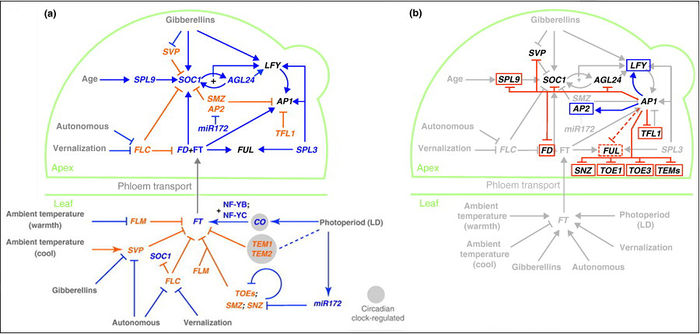
"The Arabidopsis floral transition network has been delineated as a result of extensive genetic analyses (depicted in a), but genome-wide studies have uncovered new molecular interactions and highlighted the role of AP1 as both a molecular switch and a network hub (shown in b)."
Overall pathway for the time of bloom from Wellmer and Riechmann, 2010 and Ferrier et al., 2011.
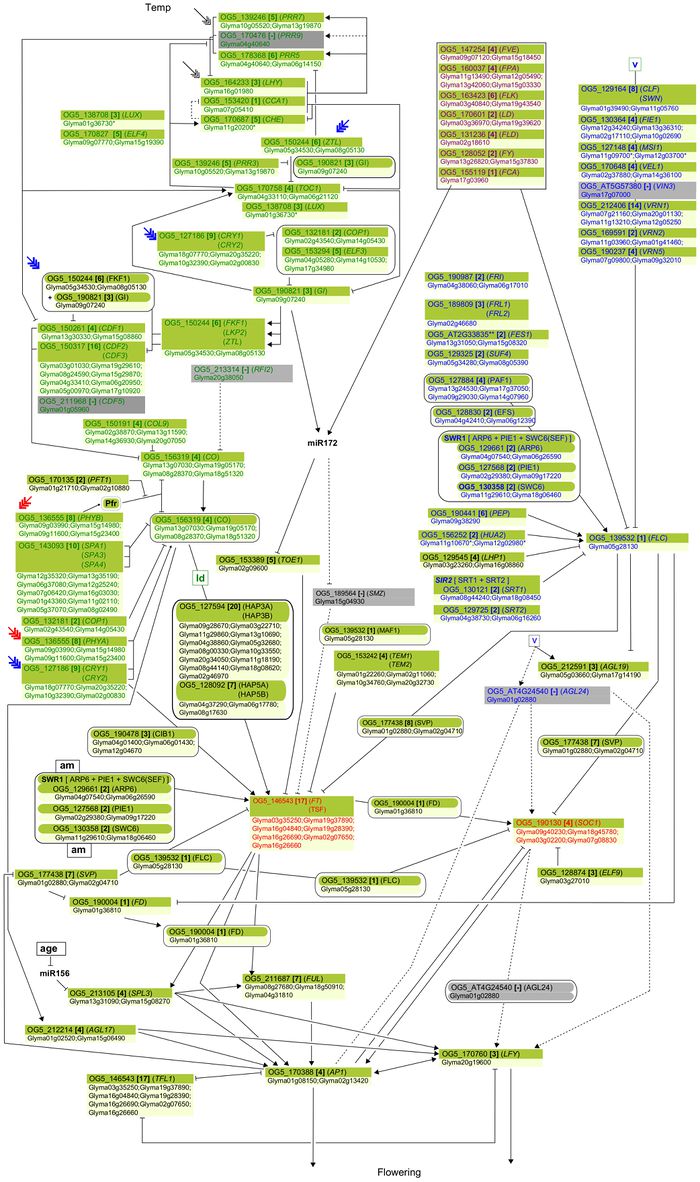
"Soybean genes marked with * are those that share the same clade with Arabidopsis genes but have not been investigated so far as flowering genes ... Arrows show promoting effects, T-bars show repressing effects. Environmental cues are shown as lower case letters in square boxes; ‘v’ is extended cold (vernalization); ‘ld’ is long days; ‘sd’ is short days; ‘am’ is ambient (non-vernalizing) temperature. Genes are shown in italics and proteins in non-italics in ovals. ‘Pfr’ indicates Pfr phytochrome signaling. Arabidopsis genes assigned to specific pathways are color-coded (photoperiod pathway in green, vernalization in blue and autonomous pathway in purple). Flowering pathway integrators are shown in red. Triple headed arrows indicate activation by red or blue light."
Overall pathway for the time of bloom from Jung et al., 2012.
All Arabidopsis Flowering Pathways
Henderson and Dean, 2004; Yamaguchi et al., 2005; Liu et al., 2009; Taiz and Zeiger, 2010; Boss et al., 2004; Zhang et al., 2011; Ballerini and Kramer, 2011; Posé et al., 2012; Kim and Sung, 2010; Wellmer and Riechmann, 2010; Farrona et al., 2008; He and Amasino, 2005; Max Planck Institute for Plant Breeding Research; Bäurle and Dean, 2006; Sung et al., 2003; Schneitz, 2009; Amasino, 2005; Ferrier et al., 2011; Izawa et al., 2003; Jung et al., 2012
Arabidopsis Flowering Genes
Note: All genes are compiled from Jung et al., 2012. Formatting is the same as Jung et al., 2012 where a * indicates 1 of "24 Arabidopsis genes, which had not been previously investigated for their roles in flowering, [that] belong to [ortholog groups] with known Arabidopsis flowering genes." Moreover, for pathways, "Am: Ambient temperature pathway, Au: Autonomous pathway, FPI: Flowering Pathway Intergrators, L: Light signaling pathway, MI: Meristem Identity, V: Vernalization pathway."
| Arabidopsis Genes | Function | Function Source / AA Sequence | Pathway |
|---|---|---|---|
| ABF1 | Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression. | UniProtKB | - |
| ABF2 | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | - |
| ABF3 | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. | UniProtKB | - |
| ABF4 | Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | - |
| ABI5 | Participates in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth. Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter and to the ABRE of the Em1 and Em6 genes promoters. Can also trans-activate its own promoter, suggesting that it is autoregulated. Plays a role in sugar-mediated senescence. | UniProtKB | - |
| AG | Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development. | UniProtKB | - |
| AGL14 | Probable transcription factor. | UniProtKB | V |
| AGL15 | Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro. | UniProtKB | - |
| AGL17 | Probable transcription factor. | UniProtKB | - |
| AGL18 | Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation. | UniProtKB | - |
| AGL19 | Probable transcription factor that promotes flowering, especially in response to vernalization by short periods of cold, in an FLC-inpedendent manner. | UniProtKB | V |
| AGL21* | Probable transcription factor. | UniProtKB | - |
| AGL24 | Transcription activator that mediates floral transition in response to vernalization. Promotes inflorescence fate in apical meristems. Acts in a dosage-dependent manner. Probably involved in the transduction of RLK-mediated signaling (e.g. IMK3 pathway). Together with AP1 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. When associated with SOC1, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. Confers inflorescence characteristics to floral primordia and early flowering. | UniProtKB | V |
| AGL31 | Probable transcription factor that prevents vernalization by short periods of cold. Acts as a floral repressor. | UniProtKB | Am, FPI, V |
| AGL6 | Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development. | UniProtKB | - |
| AGL79 | - | UniProtKB | - |
| AP1 | Transcription factor that promotes early floral meristem identity in synergy with LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Is indispensable for normal development of sepals and petals in flowers. Regulates positively the B class homeotic proteins APETALA3 and PISTILLATA with the cooperation of LEAFY and UFO. Interacts with SEPALLATA3 or AP3/PI heterodimer to form complexes that could be involved in genes regulation during floral meristem development. Regulates positively AGAMOUS in cooperation with LEAFY. Displays a redundant function with CAULIFLOWER in the up-regulation of LEAFY. Together with AGL24 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Represses flowering time genes AGL24, SVP and SOC1 in emerging floral meristems. | UniProtKB | FPI, MI |
| AP2 | Probable transcriptional activator that promotes early floral meristem identity. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Plays a central role in the specification of floral identity, particularly for the normal development of sepals and petals in the wild-type flower. Acts as A class cadastral protein by repressing the C class floral homeotic gene AGAMOUS in association with other repressors like LEUNIG and SEUSS. It is also required during seed development. | UniProtKB | - |
| AP3 | Probable transcription factor involved in the genetic control of flower development. Is required for normal development of petals and stamens in the wild-type flower. Forms a heterodimer with PISTILLATA that is required for autoregulation of both AP3 and PI genes. AP3/PI heterodimer interacts with APETALA1 or SEPALLATA3 to form a ternary complex that could be responsible for the regulation of the genes involved in the flower development. AP3/PI heterodimer activates the expression of NAP. | UniProtKB | - |
| APRR3 | Controls photoperiodic flowering response. Component of the circadian clock. Controls the degradation of APRR1/TOC1 by the SCF(ZTL) complex. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| APRR5 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| APRR9 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L |
| AREB3 | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | UniProtKB | - |