Difference between revisions of "Time of bloom"
m (→Arabidopsis Flowering Genes) |
|||
| Line 33: | Line 33: | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| − | ! <i>Arabidopsis</i> Gene !! Function (All function information is directly copied from the respective AA Sequence page) !! AA Sequence !! Pathway | + | ! <i>Arabidopsis</i> Gene !! Function (All function information is directly copied from the respective AA Sequence page) !! AA Sequence !! Pathway !! Potential Ortholog |
|- | |- | ||
| − | | ABF1||Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression.||[http://www.uniprot.org/uniprot/Q9M7Q5 UniProtKB]|| | + | | ABF1||Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression.||[http://www.uniprot.org/uniprot/Q9M7Q5 UniProtKB]|||| |
|- | |- | ||
| − | | ABF2||Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter.||[http://www.uniprot.org/uniprot/Q9M7Q4 UniProtKB]|| | + | | ABF2||Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter.||[http://www.uniprot.org/uniprot/Q9M7Q4 UniProtKB]|||| |
|- | |- | ||
| − | | ABF3||Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling.||[http://www.uniprot.org/uniprot/Q9M7Q3 UniProtKB]|| | + | | ABF3||Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling.||[http://www.uniprot.org/uniprot/Q9M7Q3 UniProtKB]|||| |
|- | |- | ||
| − | | ABF4||Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter.||[http://www.uniprot.org/uniprot/Q9M7Q2 UniProtKB]|| | + | | ABF4||Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter.||[http://www.uniprot.org/uniprot/Q9M7Q2 UniProtKB]|||| |
|- | |- | ||
| − | | ABI5||Participates in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth. Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter and to the ABRE of the Em1 and Em6 genes promoters. Can also trans-activate its own promoter, suggesting that it is autoregulated. Plays a role in sugar-mediated senescence.||[http://www.uniprot.org/uniprot/Q9SJN0 UniProtKB]|| | + | | ABI5||Participates in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth. Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter and to the ABRE of the Em1 and Em6 genes promoters. Can also trans-activate its own promoter, suggesting that it is autoregulated. Plays a role in sugar-mediated senescence.||[http://www.uniprot.org/uniprot/Q9SJN0 UniProtKB]|||| |
|- | |- | ||
| − | | AG||Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development.||[http://www.uniprot.org/uniprot/P17839 UniProtKB]|| | + | | AG||Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development.||[http://www.uniprot.org/uniprot/P17839 UniProtKB]||||[http://www.uniprot.org/uniprot/D1MDP5 Grape] |
|- | |- | ||
| − | | AGL14||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q38838 UniProtKB]||V | + | | AGL14||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q38838 UniProtKB]||V|| |
|- | |- | ||
| − | | AGL15||Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro.||[http://www.uniprot.org/uniprot/Q38847 UniProtKB]|| | + | | AGL15||Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro.||[http://www.uniprot.org/uniprot/Q38847 UniProtKB]|||| |
|- | |- | ||
| − | | AGL17||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q38840 UniProtKB]|| | + | | AGL17||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q38840 UniProtKB]|||| |
|- | |- | ||
| − | | AGL18||Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation.||[http://www.uniprot.org/uniprot/Q9M2K8 UniProtKB]|| | + | | AGL18||Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation.||[http://www.uniprot.org/uniprot/Q9M2K8 UniProtKB]|||| |
|- | |- | ||
| − | | AGL19||Probable transcription factor that promotes flowering, especially in response to vernalization by short periods of cold, in an FLC-inpedendent manner.||[http://www.uniprot.org/uniprot/O82743 UniProtKB]||V | + | | AGL19||Probable transcription factor that promotes flowering, especially in response to vernalization by short periods of cold, in an FLC-inpedendent manner.||[http://www.uniprot.org/uniprot/O82743 UniProtKB]||V|| |
|- | |- | ||
| − | | AGL21*||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q9SZJ6 UniProtKB]|| | + | | AGL21*||Probable transcription factor.||[http://www.uniprot.org/uniprot/Q9SZJ6 UniProtKB]|||| |
|- | |- | ||
| − | | AGL24||Transcription activator that mediates floral transition in response to vernalization. Promotes inflorescence fate in apical meristems. Acts in a dosage-dependent manner. Probably involved in the transduction of RLK-mediated signaling (e.g. IMK3 pathway). Together with AP1 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. When associated with SOC1, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. Confers inflorescence characteristics to floral primordia and early flowering.||[http://www.uniprot.org/uniprot/O82794 UniProtKB]||V | + | | AGL24||Transcription activator that mediates floral transition in response to vernalization. Promotes inflorescence fate in apical meristems. Acts in a dosage-dependent manner. Probably involved in the transduction of RLK-mediated signaling (e.g. IMK3 pathway). Together with AP1 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. When associated with SOC1, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. Confers inflorescence characteristics to floral primordia and early flowering.||[http://www.uniprot.org/uniprot/O82794 UniProtKB]||V|| |
|- | |- | ||
| − | | AGL31||Probable transcription factor that prevents vernalization by short periods of cold. Acts as a floral repressor.||[http://www.uniprot.org/uniprot/Q9FPN7 UniProtKB]||Am, FPI, V | + | | AGL31||Probable transcription factor that prevents vernalization by short periods of cold. Acts as a floral repressor.||[http://www.uniprot.org/uniprot/Q9FPN7 UniProtKB]||Am, FPI, V|| |
|- | |- | ||
| − | | AGL6||Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development.||[http://www.uniprot.org/uniprot/P29386 UniProtKB]|| | + | | AGL6||Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development.||[http://www.uniprot.org/uniprot/P29386 UniProtKB]|||| |
|- | |- | ||
| − | | AGL79||-||[http://www.uniprot.org/uniprot/Q7X9H6 UniProtKB]|| | + | | AGL79||-||[http://www.uniprot.org/uniprot/Q7X9H6 UniProtKB]|||| |
|- | |- | ||
| − | | AP1||Transcription factor that promotes early floral meristem identity in synergy with LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Is indispensable for normal development of sepals and petals in flowers. Regulates positively the B class homeotic proteins APETALA3 and PISTILLATA with the cooperation of LEAFY and UFO. Interacts with SEPALLATA3 or AP3/PI heterodimer to form complexes that could be involved in genes regulation during floral meristem development. Regulates positively AGAMOUS in cooperation with LEAFY. Displays a redundant function with CAULIFLOWER in the up-regulation of LEAFY. Together with AGL24 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Represses flowering time genes AGL24, SVP and SOC1 in emerging floral meristems.||[http://www.uniprot.org/uniprot/P35631 UniProtKB]||FPI, MI | + | | AP1||Transcription factor that promotes early floral meristem identity in synergy with LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Is indispensable for normal development of sepals and petals in flowers. Regulates positively the B class homeotic proteins APETALA3 and PISTILLATA with the cooperation of LEAFY and UFO. Interacts with SEPALLATA3 or AP3/PI heterodimer to form complexes that could be involved in genes regulation during floral meristem development. Regulates positively AGAMOUS in cooperation with LEAFY. Displays a redundant function with CAULIFLOWER in the up-regulation of LEAFY. Together with AGL24 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Represses flowering time genes AGL24, SVP and SOC1 in emerging floral meristems.||[http://www.uniprot.org/uniprot/P35631 UniProtKB]||FPI, MI||[http://www.uniprot.org/uniprot/D1MDP8 Grape] |
|- | |- | ||
| − | | AP2||Probable transcriptional activator that promotes early floral meristem identity. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Plays a central role in the specification of floral identity, particularly for the normal development of sepals and petals in the wild-type flower. Acts as A class cadastral protein by repressing the C class floral homeotic gene AGAMOUS in association with other repressors like LEUNIG and SEUSS. It is also required during seed development.||[http://www.uniprot.org/uniprot/P47927 UniProtKB]|| | + | | AP2||Probable transcriptional activator that promotes early floral meristem identity. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Plays a central role in the specification of floral identity, particularly for the normal development of sepals and petals in the wild-type flower. Acts as A class cadastral protein by repressing the C class floral homeotic gene AGAMOUS in association with other repressors like LEUNIG and SEUSS. It is also required during seed development.||[http://www.uniprot.org/uniprot/P47927 UniProtKB]||||[http://www.uniprot.org/uniprot/C1KBJ2 Grape] |
|- | |- | ||
| − | | AP3||Probable transcription factor involved in the genetic control of flower development. Is required for normal development of petals and stamens in the wild-type flower. Forms a heterodimer with PISTILLATA that is required for autoregulation of both AP3 and PI genes. AP3/PI heterodimer interacts with APETALA1 or SEPALLATA3 to form a ternary complex that could be responsible for the regulation of the genes involved in the flower development. AP3/PI heterodimer activates the expression of NAP.||[http://www.uniprot.org/uniprot/P35632 UniProtKB]|| | + | | AP3||Probable transcription factor involved in the genetic control of flower development. Is required for normal development of petals and stamens in the wild-type flower. Forms a heterodimer with PISTILLATA that is required for autoregulation of both AP3 and PI genes. AP3/PI heterodimer interacts with APETALA1 or SEPALLATA3 to form a ternary complex that could be responsible for the regulation of the genes involved in the flower development. AP3/PI heterodimer activates the expression of NAP.||[http://www.uniprot.org/uniprot/P35632 UniProtKB]||||[http://www.uniprot.org/uniprot/A3RJI1 Grape] |
|- | |- | ||
| − | | APRR3||Controls photoperiodic flowering response. Component of the circadian clock. Controls the degradation of APRR1/TOC1 by the SCF(ZTL) complex. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q9LVG4 UniProtKB]||L | + | | APRR3||Controls photoperiodic flowering response. Component of the circadian clock. Controls the degradation of APRR1/TOC1 by the SCF(ZTL) complex. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q9LVG4 UniProtKB]||L|| |
|- | |- | ||
| − | | APRR5||Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q6LA42 UniProtKB]||L | + | | APRR5||Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q6LA42 UniProtKB]||L|| |
|- | |- | ||
| − | | APRR9||Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q8L500 UniProtKB]||L | + | | APRR9||Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock.||[http://www.uniprot.org/uniprot/Q8L500 UniProtKB]||L|| |
|- | |- | ||
| − | | AREB3||Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development.||[http://www.uniprot.org/uniprot/Q9LES3 UniProtKB]|| | + | | AREB3||Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development.||[http://www.uniprot.org/uniprot/Q9LES3 UniProtKB]|||| |
|- | |- | ||
| − | | AT1G26790||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence.||[http://www.uniprot.org/uniprot/Q9LQX4 UniProtKB]||L | + | | AT1G26790||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence.||[http://www.uniprot.org/uniprot/Q9LQX4 UniProtKB]||L|| |
|- | |- | ||
| − | | AT1G29160||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Acts as a negative regulator in the phytochrome-mediated light responses. Controls phyB-mediated end-of-day response and the phyA-mediated anthocyanin accumulation.||[http://www.uniprot.org/uniprot/P68350 UniProtKB]||L | + | | AT1G29160||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Acts as a negative regulator in the phytochrome-mediated light responses. Controls phyB-mediated end-of-day response and the phyA-mediated anthocyanin accumulation.||[http://www.uniprot.org/uniprot/P68350 UniProtKB]||L|| |
|- | |- | ||
| − | | AT1G50680||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9C6P5 UniProtKB]|| | + | | AT1G50680||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9C6P5 UniProtKB]|||| |
|- | |- | ||
| − | | AT1G51120||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9C688 UniProtKB]|| | + | | AT1G51120||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9C688 UniProtKB]|||| |
|- | |- | ||
| − | | AT2G23070||Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins.||[http://www.uniprot.org/uniprot/O64816 UniProtKB]|| | + | | AT2G23070||Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins.||[http://www.uniprot.org/uniprot/O64816 UniProtKB]|||| |
|- | |- | ||
| − | | AT2G23080||Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins.||[http://www.uniprot.org/uniprot/O64817 UniProtKB]|| | + | | AT2G23080||Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins.||[http://www.uniprot.org/uniprot/O64817 UniProtKB]|||| |
|- | |- | ||
| − | | AT2G25920||Uncharacterized protein||[http://www.uniprot.org/uniprot/O82305 UniProtKB]|| | + | | AT2G25920||Uncharacterized protein||[http://www.uniprot.org/uniprot/O82305 UniProtKB]|||| |
|- | |- | ||
| − | | AT2G34140||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence.||[http://www.uniprot.org/uniprot/O22967 UniProtKB]||L | + | | AT2G34140||Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence.||[http://www.uniprot.org/uniprot/O22967 UniProtKB]||L|| |
|- | |- | ||
| − | | AT2G46670*||Putative uncharacterized protein||[http://www.uniprot.org/uniprot/Q683A6 UniProtKB]||L | + | | AT2G46670*||Putative uncharacterized protein||[http://www.uniprot.org/uniprot/Q683A6 UniProtKB]||L|| |
|- | |- | ||
| − | | AT3G21320||Uncharacterized protein||[http://www.uniprot.org/uniprot/Q5Q0C8 UniProtKB]||L | + | | AT3G21320||Uncharacterized protein||[http://www.uniprot.org/uniprot/Q5Q0C8 UniProtKB]||L|| |
|- | |- | ||
| − | | AT3G25730||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9LS06 UniProtKB]||L | + | | AT3G25730||Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways.||[http://www.uniprot.org/uniprot/Q9LS06 UniProtKB]||L|| |
|- | |- | ||
| − | | AT4G16810||Putative uncharacterized protein||[http://www.uniprot.org/uniprot/O23521 UniProtKB]|| | + | | AT4G16810||Putative uncharacterized protein||[http://www.uniprot.org/uniprot/O23521 UniProtKB]|||| |
|- | |- | ||
| − | | AT5G08230*||Probable transcrition factor that seems to be involved in mRNA processing.||[http://www.uniprot.org/uniprot/Q9LEY4 UniProtKB]||V | + | | AT5G08230*||Probable transcrition factor that seems to be involved in mRNA processing.||[http://www.uniprot.org/uniprot/Q9LEY4 UniProtKB]||V|| |
|- | |- | ||
| − | | AT5G10625*||Modulates the competence to flowering of apical meristems.||[http://www.uniprot.org/uniprot/Q9LXB5 UniProtKB]|| | + | | AT5G10625*||Modulates the competence to flowering of apical meristems.||[http://www.uniprot.org/uniprot/Q9LXB5 UniProtKB]|||| |
|- | |- | ||
| − | | AT5G23280*||Transcription factor||[http://www.uniprot.org/uniprot/Q9FMX2 UniProtKB]||L | + | | AT5G23280*||Transcription factor||[http://www.uniprot.org/uniprot/Q9FMX2 UniProtKB]||L|| |
|- | |- | ||
| − | | AT5G27220||Frigida-like protein||[http://www.uniprot.org/uniprot/O04650 UniProtKB]|| | + | | AT5G27220||Frigida-like protein||[http://www.uniprot.org/uniprot/O04650 UniProtKB]|||| |
|- | |- | ||
| − | | AT5G42910||Could participate in abscisic acid-regulated gene expression.||[http://www.uniprot.org/uniprot/Q9FMM7 UniProtKB]|| | + | | AT5G42910||Could participate in abscisic acid-regulated gene expression.||[http://www.uniprot.org/uniprot/Q9FMM7 UniProtKB]|||| |
|- | |- | ||
| − | | AT5G44080||Basic leucine zipper transcription factor||[http://www.uniprot.org/uniprot/Q9FNB9 UniProtKB]|| | + | | AT5G44080||Basic leucine zipper transcription factor||[http://www.uniprot.org/uniprot/Q9FNB9 UniProtKB]|||| |
|- | |- | ||
| − | | AT5G48250*||Zinc finger protein Constans-like 10||[http://www.uniprot.org/uniprot/Q9LUA9 UniProtKB]||L | + | | AT5G48250*||Zinc finger protein Constans-like 10||[http://www.uniprot.org/uniprot/Q9LUA9 UniProtKB]||L|| |
|- | |- | ||
| − | | AT5G59570*||Putative transcription factor||[http://www.uniprot.org/uniprot/Q9LTH4 UniProtKB]||L | + | | AT5G59570*||Putative transcription factor||[http://www.uniprot.org/uniprot/Q9LTH4 UniProtKB]||L|| |
|- | |- | ||
| − | | AT5G62040||May form complexes with phosphorylated ligands by interfering with kinases and their effectors.||[http://www.uniprot.org/uniprot/Q9FIT4 UniProtKB]|| | + | | AT5G62040||May form complexes with phosphorylated ligands by interfering with kinases and their effectors.||[http://www.uniprot.org/uniprot/Q9FIT4 UniProtKB]|||| |
|- | |- | ||
| − | | ATARP6||Component of the SWR1 complex which mediates the ATP-dependent exchange of histone H2A for the H2A variant H2A.F/Z leading to transcriptional regulation of selected genes (e.g. FLC) by chromatin remodeling. Required for the activation of FLC and FLC/MAF genes expression to levels that inhibit flowering, through both histone H3 and H4 acetylation and methylation mechanisms. Involved in several developmental processes including organization of plant organs, leaves formation, flowering time repression, and fertility. Modulates photoperiod-dependent epidermal leaves cell development; promotes cell division in long days, and cell expansion/division in short days. May be involved in the regulation of pathogenesis-related proteins (PRs).||[http://www.uniprot.org/uniprot/Q8LGE3 UniProtKB]||Am, V | + | | ATARP6||Component of the SWR1 complex which mediates the ATP-dependent exchange of histone H2A for the H2A variant H2A.F/Z leading to transcriptional regulation of selected genes (e.g. FLC) by chromatin remodeling. Required for the activation of FLC and FLC/MAF genes expression to levels that inhibit flowering, through both histone H3 and H4 acetylation and methylation mechanisms. Involved in several developmental processes including organization of plant organs, leaves formation, flowering time repression, and fertility. Modulates photoperiod-dependent epidermal leaves cell development; promotes cell division in long days, and cell expansion/division in short days. May be involved in the regulation of pathogenesis-related proteins (PRs).||[http://www.uniprot.org/uniprot/Q8LGE3 UniProtKB]||Am, V|| |
|- | |- | ||
| − | | ATBZIP27||Transcription factor||[http://www.genscript.com/cgi-bin/orf/refseq.pl?acc=NM_127331 GenScript]||Am, MI | + | | ATBZIP27||Transcription factor||[http://www.genscript.com/cgi-bin/orf/refseq.pl?acc=NM_127331 GenScript]||Am, MI|| |
|- | |- | ||
| − | | ATC||May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro).||[http://www.uniprot.org/uniprot/Q9ZNV5 UniProtKB]|| | + | | ATC||May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro).||[http://www.uniprot.org/uniprot/Q9ZNV5 UniProtKB]|||| |
|- | |- | ||
| − | | ATCOL4||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=131285&type=locus TAIR]||L | + | | ATCOL4||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=131285&type=locus TAIR]||L|| |
|- | |- | ||
| − | | ATCOL5||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=134422&type=locus TAIR]||L | + | | ATCOL5||Sequence-specific DNA binding transcription factor activity, zinc ion binding.||[http://arabidopsis.org/servlets/TairObject?id=134422&type=locus TAIR]||L|| |
|- | |- | ||
| − | | ATSWC6||Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes.||[http://arabidopsis.org/servlets/TairObject?id=500231974&type=locus TAIR]||V | + | | ATSWC6||Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes.||[http://arabidopsis.org/servlets/TairObject?id=500231974&type=locus TAIR]||V|| |
|- | |- | ||
| − | | CAL||Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, | + | | CAL||Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, FRUITFULL and LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Seems to be partially redundant to the function of APETALA1.||[http://www.uniprot.org/uniprot/Q39375 UniProtKB]||FPI, MI|| |
|- | |- | ||
| − | | CCA1||Transcription factor involved in the circadian clock and in the phytochrome regulation. Binds to the promoter regions of APRR1/TOC1 and TCP21/CHE to repress their transcription. Binds to the promoter regions of CAB2A and CAB2B to promote their transcription. Represses both LHY and itself.||[http://www.uniprot.org/uniprot/P92973 UniProtKB]||L | + | | CCA1||Transcription factor involved in the circadian clock and in the phytochrome regulation. Binds to the promoter regions of APRR1/TOC1 and TCP21/CHE to repress their transcription. Binds to the promoter regions of CAB2A and CAB2B to promote their transcription. Represses both LHY and itself.||[http://www.uniprot.org/uniprot/P92973 UniProtKB]||L|| |
|- | |- | ||
| − | | CDF1||Cell growth defect factor||[http://www.uniprot.org/uniprot/Q4ACT9 UniProtKB]||L | + | | CDF1||Cell growth defect factor||[http://www.uniprot.org/uniprot/Q4ACT9 UniProtKB]||L|| |
|- | |- | ||
| − | | CDF2||Cell growth defect factor||[http://www.uniprot.org/uniprot/Q3C1C7 UniProtKB]||L | + | | CDF2||Cell growth defect factor||[http://www.uniprot.org/uniprot/Q3C1C7 UniProtKB]||L|| |
|- | |- | ||
| − | | CDF3||Plant non-specific lipid-transfer proteins transfer phospholipids as well as galactolipids across membranes. May play a role in wax or cutin deposition in the cell walls of expanding epidermal cells and certain secretory tissues.||[http://www.uniprot.org/uniprot/Q9S7I3 UniProtKB]||L | + | | CDF3||Plant non-specific lipid-transfer proteins transfer phospholipids as well as galactolipids across membranes. May play a role in wax or cutin deposition in the cell walls of expanding epidermal cells and certain secretory tissues.||[http://www.uniprot.org/uniprot/Q9S7I3 UniProtKB]||L|| |
|- | |- | ||
| − | | CDF5||Cell growth defect factor||-||L | + | | CDF5||Cell growth defect factor||-||L|| |
|- | |- | ||
| − | | CHE||-||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||L | + | | CHE||-||[http://www.uniprot.org/uniprot/Q9FTA2 UniProtKB]||L|| |
|- | |- | ||
| − | | CIB1||Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression.||[http://arabidopsis.org/servlets/TairObject?id=130033&type=locus TAIR]||L | + | | CIB1||Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression.||[http://arabidopsis.org/servlets/TairObject?id=130033&type=locus TAIR]||L|| |
|- | |- | ||
| − | | CKA1||Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis.||[http://arabidopsis.org/servlets/TairObject?id=132478&type=locus TAIR]|| | + | | CKA1||Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis.||[http://arabidopsis.org/servlets/TairObject?id=132478&type=locus TAIR]|||| |
|- | |- | ||
| − | | CKA2||Encodes the casein kinase II (CK2) catalytic subunit (alpha).||[http://arabidopsis.org/servlets/TairObject?id=37139&type=locus TAIR]|| | + | | CKA2||Encodes the casein kinase II (CK2) catalytic subunit (alpha).||[http://arabidopsis.org/servlets/TairObject?id=37139&type=locus TAIR]|||| |
|- | |- | ||
| − | | CLF||Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant.||[http://arabidopsis.org/servlets/TairObject?id=26534&type=locus TAIR]||Au, V | + | | CLF||Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant.||[http://arabidopsis.org/servlets/TairObject?id=26534&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | CO||Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1.||[http://arabidopsis.org/servlets/TairObject?id=130492&type=locus TAIR]||FPI, L | + | | CO||Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1.||[http://arabidopsis.org/servlets/TairObject?id=130492&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | COL1||Homologous to the flowering-time gene CONSTANS.||[http://arabidopsis.org/servlets/TairObject?id=130495&type=locus TAIR]||FPI, L | + | | COL1||Homologous to the flowering-time gene CONSTANS.||[http://arabidopsis.org/servlets/TairObject?id=130495&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | COL2||Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins||[http://arabidopsis.org/servlets/TairObject?id=35635&type=locus TAIR]||FPI, L | + | | COL2||Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins||[http://arabidopsis.org/servlets/TairObject?id=35635&type=locus TAIR]||FPI, L|| |
|- | |- | ||
| − | | COL3||Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching.||[http://arabidopsis.org/servlets/TairObject?id=32704&type=locus TAIR]|| | + | | COL3||Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching.||[http://arabidopsis.org/servlets/TairObject?id=32704&type=locus TAIR]|||| |
|- | |- | ||
| − | | COL9||This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO.||[http://arabidopsis.org/servlets/TairObject?id=38550&type=locus TAIR]||L | + | | COL9||This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO.||[http://arabidopsis.org/servlets/TairObject?id=38550&type=locus TAIR]||L|| |
|- | |- | ||
| − | | COP1||Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light.||[http://arabidopsis.org/servlets/TairObject?id=34462&type=locus TAIR]||L | + | | COP1||Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light.||[http://arabidopsis.org/servlets/TairObject?id=34462&type=locus TAIR]||L|| |
|- | |- | ||
| − | | CRY1||Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings.||[http://arabidopsis.org/servlets/TairObject?id=129943&type=locus TAIR]||L | + | | CRY1||Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings.||[http://arabidopsis.org/servlets/TairObject?id=129943&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/B6REW9 Grape] |
|- | |- | ||
| − | | CRY2||Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus.||[http://arabidopsis.org/servlets/TairObject?id=28374&type=locus TAIR]||L | + | | CRY2||Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus.||[http://arabidopsis.org/servlets/TairObject?id=28374&type=locus TAIR]||L|| |
|- | |- | ||
| − | | DPBF2||Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons.||[http://arabidopsis.org/servlets/TairObject?id=35893&type=locus TAIR]|| | + | | DPBF2||Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons.||[http://arabidopsis.org/servlets/TairObject?id=35893&type=locus TAIR]|||| |
|- | |- | ||
| − | | E12A11||Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed.||[http://arabidopsis.org/servlets/TairObject?id=136229&type=locus TAIR]|| | + | | E12A11||Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed.||[http://arabidopsis.org/servlets/TairObject?id=136229&type=locus TAIR]|||| |
|- | |- | ||
| − | | EEL||Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo.||[http://arabidopsis.org/servlets/TairObject?id=35106&type=locus TAIR]|| | + | | EEL||Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo.||[http://arabidopsis.org/servlets/TairObject?id=35106&type=locus TAIR]|||| |
|- | |- | ||
| − | | EFS||Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development.||[http://arabidopsis.org/servlets/TairObject?id=136429&type=locus TAIR]||V | + | | EFS||Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development.||[http://arabidopsis.org/servlets/TairObject?id=136429&type=locus TAIR]||V|| |
|- | |- | ||
| − | | ELF3||Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions.||[http://arabidopsis.org/servlets/TairObject?id=32082&type=locus TAIR]||L | + | | ELF3||Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions.||[http://arabidopsis.org/servlets/TairObject?id=32082&type=locus TAIR]||L|| |
|- | |- | ||
| − | | ELF4||Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes.||[http://arabidopsis.org/servlets/TairObject?id=34766&type=locus TAIR]||L | + | | ELF4||Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes.||[http://arabidopsis.org/servlets/TairObject?id=34766&type=locus TAIR]||L|| |
|- | |- | ||
| − | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32448&type=locus TAIR]|| | + | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32448&type=locus TAIR]|||| |
|- | |- | ||
| − | | ELF4-L2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=29911&type=locus TAIR]|| | + | | ELF4-L2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=29911&type=locus TAIR]|||| |
|- | |- | ||
| − | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231553&type=locus TAIR]||L | + | | ELF4-L3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231553&type=locus TAIR]||L|| |
|- | |- | ||
| − | | ELF4-L4||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231439&type=locus TAIR]|| | + | | ELF4-L4||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=500231439&type=locus TAIR]|||| |
|- | |- | ||
| − | | ELF5||Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC||[http://arabidopsis.org/servlets/TairObject?id=134377&type=locus TAIR]|| | + | | ELF5||Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC||[http://arabidopsis.org/servlets/TairObject?id=134377&type=locus TAIR]|||| |
|- | |- | ||
| − | | ELF6||Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=130928&type=locus TAIR]|| | + | | ELF6||Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=130928&type=locus TAIR]|||| |
|- | |- | ||
| − | | ELF9||Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay.||[http://arabidopsis.org/servlets/TairObject?id=135649&type=locus TAIR]||FPI | + | | ELF9||Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay.||[http://arabidopsis.org/servlets/TairObject?id=135649&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | EMF1||Involved in regulating reproductive development||[http://arabidopsis.org/servlets/TairObject?id=130620&type=locus TAIR]|| | + | | EMF1||Involved in regulating reproductive development||[http://arabidopsis.org/servlets/TairObject?id=130620&type=locus TAIR]|||| |
|- | |- | ||
| − | | EMF2||Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3.||[http://arabidopsis.org/servlets/TairObject?id=134952&type=locus TAIR]|| | + | | EMF2||Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3.||[http://arabidopsis.org/servlets/TairObject?id=134952&type=locus TAIR]|||| |
|- | |- | ||
| − | | ESD4||EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development.||[http://arabidopsis.org/servlets/TairObject?id=128953&type=locus TAIR]|| | + | | ESD4||EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development.||[http://arabidopsis.org/servlets/TairObject?id=128953&type=locus TAIR]|||| |
|- | |- | ||
| − | | FCA||Involved in the promotion of the transition of the vegetative meristem to reproductive development. Four forms of the protein (alpha, beta, delta and gamma) are produced by alternative splicing. Involved in RNA-mediated chromatin silencing. At one point it was believed to act as an abscisic acid receptor but the paper describing that function was retracted.||[http://arabidopsis.org/servlets/TairObject?id=128853&type=locus TAIR]||Am, Au | + | | FCA||Involved in the promotion of the transition of the vegetative meristem to reproductive development. Four forms of the protein (alpha, beta, delta and gamma) are produced by alternative splicing. Involved in RNA-mediated chromatin silencing. At one point it was believed to act as an abscisic acid receptor but the paper describing that function was retracted.||[http://arabidopsis.org/servlets/TairObject?id=128853&type=locus TAIR]||Am, Au||[http://www.uniprot.org/uniprot/D2Y3W8 Grape] |
|- | |- | ||
| − | | FD||bZIP protein required for positive regulation of flowering. Mutants are late flowering. FD interacts with FT to promote flowering.Expressed in the shoot apex in floral anlagen, then declines in floral primordia.||[http://arabidopsis.org/servlets/TairObject?id=128068&type=locus TAIR]||Am, MI | + | | FD||bZIP protein required for positive regulation of flowering. Mutants are late flowering. FD interacts with FT to promote flowering.Expressed in the shoot apex in floral anlagen, then declines in floral primordia.||[http://arabidopsis.org/servlets/TairObject?id=128068&type=locus TAIR]||Am, MI|| |
|- | |- | ||
| − | | FES1||Encodes a zinc finger domain containing protein that is expressed in the shoot/root apex and vasculature, and acts with FRI to repress flowering.FES1 mutants in a Col(FRI+) background will flower early under inductive conditions.||[http://arabidopsis.org/servlets/TairObject?id=500230872&type=locus TAIR]||V | + | | FES1||Encodes a zinc finger domain containing protein that is expressed in the shoot/root apex and vasculature, and acts with FRI to repress flowering.FES1 mutants in a Col(FRI+) background will flower early under inductive conditions.||[http://arabidopsis.org/servlets/TairObject?id=500230872&type=locus TAIR]||V|| |
|- | |- | ||
| − | | FIE1||Encodes a protein similar to the transcriptional regular of the animal Polycomb group and is involved in regulation of establishment of anterior-posterior polar axis in the endosperm and repression of flowering during vegetative phase. Mutation leads endosperm to develop in the absence of fertilization and flowers to form in seedlings and non-reproductive organs. Also exhibits maternal effect gametophytic lethal phenotype, which is suppressed by hypomethylation. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. In the ovule, the FIE transcript levels increase transiently just after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=38691&type=locus TAIR]||Au, V | + | | FIE1||Encodes a protein similar to the transcriptional regular of the animal Polycomb group and is involved in regulation of establishment of anterior-posterior polar axis in the endosperm and repression of flowering during vegetative phase. Mutation leads endosperm to develop in the absence of fertilization and flowers to form in seedlings and non-reproductive organs. Also exhibits maternal effect gametophytic lethal phenotype, which is suppressed by hypomethylation. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. In the ovule, the FIE transcript levels increase transiently just after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=38691&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | FIS2||Encodes a negative regulator of seed development in the absence of pollination. In the ovule, the FIS2 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=26543&type=locus TAIR]|| | + | | FIS2||Encodes a negative regulator of seed development in the absence of pollination. In the ovule, the FIS2 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization.||[http://arabidopsis.org/servlets/TairObject?id=26543&type=locus TAIR]|||| |
|- | |- | ||
| − | | FKF1||Encodes FKF1, a flavin-binding kelch repeat F box protein, is clock-controlled, regulates transition to flowering. Forms a complex with GI on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=137051&type=locus TAIR]||L | + | | FKF1||Encodes FKF1, a flavin-binding kelch repeat F box protein, is clock-controlled, regulates transition to flowering. Forms a complex with GI on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=137051&type=locus TAIR]||L|| |
|- | |- | ||
| − | | FLC||MADS-box protein encoded by FLOWERING LOCUS C - transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock. Expression is downregulated during cold treatment. Vernalization, FRI and the autonomous pathway all influence the state of FLC chromatin. Both maternal and paternal alleles are reset by vernalization, but their earliest activation differs in timing and location. Histone H3 trimethylation at lysine 4 and histone acetylation are associated with active FLC expression, whereas histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC repression. Expression is also repressed by two small RNAs (30- and 24-nt) complementary to the FLC sense strand 3’ to the polyA site. The small RNAs are most likely derived from an antisense transcript of FLC. Interacts with SOC1 and FT chromatin in vivo. Member of a protein complex.||[http://arabidopsis.org/servlets/TairObject?id=136002&type=locus TAIR]||Am, FPI, V | + | | FLC||MADS-box protein encoded by FLOWERING LOCUS C - transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock. Expression is downregulated during cold treatment. Vernalization, FRI and the autonomous pathway all influence the state of FLC chromatin. Both maternal and paternal alleles are reset by vernalization, but their earliest activation differs in timing and location. Histone H3 trimethylation at lysine 4 and histone acetylation are associated with active FLC expression, whereas histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC repression. Expression is also repressed by two small RNAs (30- and 24-nt) complementary to the FLC sense strand 3’ to the polyA site. The small RNAs are most likely derived from an antisense transcript of FLC. Interacts with SOC1 and FT chromatin in vivo. Member of a protein complex.||[http://arabidopsis.org/servlets/TairObject?id=136002&type=locus TAIR]||Am, FPI, V||[http://www.uniprot.org/uniprot/D1MDP4 Grape] |
|- | |- | ||
| − | | FLD||Encodes a plant homolog of a SWIRM domain containing protein found in histone deacetylase complexes in mammals. Lesions in FLD result in hyperacetylation of histones in FLC chromatin, up-regulation of FLC expression and extremely delayed flowering. FLD plays a key role in regulating the reproductive competence of the shoot and results in different developmental phase transitions in Arabidopsis.||[http://arabidopsis.org/servlets/TairObject?id=35869&type=locus TAIR]||Am, Au | + | | FLD||Encodes a plant homolog of a SWIRM domain containing protein found in histone deacetylase complexes in mammals. Lesions in FLD result in hyperacetylation of histones in FLC chromatin, up-regulation of FLC expression and extremely delayed flowering. FLD plays a key role in regulating the reproductive competence of the shoot and results in different developmental phase transitions in Arabidopsis.||[http://arabidopsis.org/servlets/TairObject?id=35869&type=locus TAIR]||Am, Au|| |
|- | |- | ||
| − | | FLK||RNA binding, nucleic acid binding; positive regulation of flower development||[http://arabidopsis.org/servlets/TairObject?id=37455&type=locus TAIR]||Au | + | | FLK||RNA binding, nucleic acid binding; positive regulation of flower development||[http://arabidopsis.org/servlets/TairObject?id=37455&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | FLP1*||Encodes a small protein with unknown function and is similar to flower promoting factor 1. This gene is not expressed in apical meristem after floral induction but is expressed in roots, flowers, and in low abundance, leaves.||[http://arabidopsis.org/servlets/TairObject?id=128483&type=locus TAIR]|| | + | | FLP1*||Encodes a small protein with unknown function and is similar to flower promoting factor 1. This gene is not expressed in apical meristem after floral induction but is expressed in roots, flowers, and in low abundance, leaves.||[http://arabidopsis.org/servlets/TairObject?id=128483&type=locus TAIR]|||| |
|- | |- | ||
| − | | FPA||FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. Double mutants with FCA have reduced fertility and single/double mutants have defects in siRNA mediated chromatin silencing.||[http://arabidopsis.org/servlets/TairObject?id=34279&type=locus TAIR]||Au | + | | FPA||FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. Double mutants with FCA have reduced fertility and single/double mutants have defects in siRNA mediated chromatin silencing.||[http://arabidopsis.org/servlets/TairObject?id=34279&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | FPF1||Encodes a small protein of 12.6 kDa that regulates flowering and is involved in gibberellin signalling pathway. It is expressed in apical meristems immediately after the photoperiodic induction of flowering. Genetic interactions with flowering time and floral organ identity genes suggest that this gene may be involved in modulating the competence to flower. There are two other genes similar to FPF1, FLP1 (At4g31380) and FLP2 (no locus name yet, on BAC F8F16 on chr 4). This is so far a plant-specific gene and is only found in long-day mustard, arabidopsis, and rice.||[http://arabidopsis.org/servlets/TairObject?id=131288&type=locus TAIR]|| | + | | FPF1||Encodes a small protein of 12.6 kDa that regulates flowering and is involved in gibberellin signalling pathway. It is expressed in apical meristems immediately after the photoperiodic induction of flowering. Genetic interactions with flowering time and floral organ identity genes suggest that this gene may be involved in modulating the competence to flower. There are two other genes similar to FPF1, FLP1 (At4g31380) and FLP2 (no locus name yet, on BAC F8F16 on chr 4). This is so far a plant-specific gene and is only found in long-day mustard, arabidopsis, and rice.||[http://arabidopsis.org/servlets/TairObject?id=131288&type=locus TAIR]|||| |
|- | |- | ||
| − | | FRI||Encodes a major determinant of natural variation in Arabidopsis flowering time. Dominant alleles of FRI confer a vernalization requirement causing plants to overwinter vegetatively. Many early flowering accessions carry loss-of-function fri alleles .Twenty distinct haplotypes that contain non-functional FRI alleles have been identified and the distribution analyzed in over 190 accessions. The common lab strains- Col and Ler each carry loss of function mutations in FRI.||[http://arabidopsis.org/servlets/TairObject?id=128299&type=locus TAIR]||V | + | | FRI||Encodes a major determinant of natural variation in Arabidopsis flowering time. Dominant alleles of FRI confer a vernalization requirement causing plants to overwinter vegetatively. Many early flowering accessions carry loss-of-function fri alleles .Twenty distinct haplotypes that contain non-functional FRI alleles have been identified and the distribution analyzed in over 190 accessions. The common lab strains- Col and Ler each carry loss of function mutations in FRI.||[http://arabidopsis.org/servlets/TairObject?id=128299&type=locus TAIR]||V|| |
|- | |- | ||
| − | | FRL1||Encodes a sterol-C24-methyltransferases involved in sterol biosynthesis. Mutants display altered sterol composition, serrated petals and sepals and altered cotyledon vascular patterning as well as ectopic endoreduplication. This suggests that suppression of endoreduplication is important for petal morphogenesis and that normal sterol composition is required for this suppression.||[http://arabidopsis.org/servlets/TairObject?id=27665&type=locus TAIR]||V | + | | FRL1||Encodes a sterol-C24-methyltransferases involved in sterol biosynthesis. Mutants display altered sterol composition, serrated petals and sepals and altered cotyledon vascular patterning as well as ectopic endoreduplication. This suggests that suppression of endoreduplication is important for petal morphogenesis and that normal sterol composition is required for this suppression.||[http://arabidopsis.org/servlets/TairObject?id=27665&type=locus TAIR]||V|| |
|- | |- | ||
| − | | FRL2||Family member of FRI-related genes that is required for the winter-annual habit. ||[http://arabidopsis.org/servlets/TairObject?id=226970&type=locus TAIR]||V | + | | FRL2||Family member of FRI-related genes that is required for the winter-annual habit. ||[http://arabidopsis.org/servlets/TairObject?id=226970&type=locus TAIR]||V|| |
|- | |- | ||
| − | | FT||FT, together with LFY, promotes flowering and is antagonistic with its homologous gene, TERMINAL FLOWER1 (TFL1). FT is expressed in leaves and is induced by long day treatment. Either the FT mRNA or protein is translocated to the shoot apex where it induces its own expression. Recent data suggests that FT protein acts as a long-range signal. FT is a target of CO and acts upstream of SOC1.||[http://arabidopsis.org/servlets/TairObject?id=30541&type=locus TAIR]||Am, FPI | + | | FT||FT, together with LFY, promotes flowering and is antagonistic with its homologous gene, TERMINAL FLOWER1 (TFL1). FT is expressed in leaves and is induced by long day treatment. Either the FT mRNA or protein is translocated to the shoot apex where it induces its own expression. Recent data suggests that FT protein acts as a long-range signal. FT is a target of CO and acts upstream of SOC1.||[http://arabidopsis.org/servlets/TairObject?id=30541&type=locus TAIR]||Am, FPI||[http://www.uniprot.org/uniprot/D1MDP3 Grape] |
|- | |- | ||
| − | | FUL||MADS box gene negatively regulated by APETALA1||[http://arabidopsis.org/servlets/TairObject?id=134558&type=locus TAIR]||MI | + | | FUL||MADS box gene negatively regulated by APETALA1||[http://arabidopsis.org/servlets/TairObject?id=134558&type=locus TAIR]||MI||[http://www.uniprot.org/uniprot/D1MDP9 Grape] |
|- | |- | ||
| − | | FVE||Controls flowering.||[http://arabidopsis.org/servlets/TairObject?id=33019&type=locus TAIR]||Am, Au | + | | FVE||Controls flowering.||[http://arabidopsis.org/servlets/TairObject?id=33019&type=locus TAIR]||Am, Au|| |
|- | |- | ||
| − | | FY||Encodes a protein with similarity to yeast Pfs2p, an mRNA processing factor. Involved in regulation of flowering time; affects FCA mRNA processing. Homozygous mutants are late flowering, null alleles are embryo lethal.||[http://arabidopsis.org/servlets/TairObject?id=136108&type=locus TAIR]||Au | + | | FY||Encodes a protein with similarity to yeast Pfs2p, an mRNA processing factor. Involved in regulation of flowering time; affects FCA mRNA processing. Homozygous mutants are late flowering, null alleles are embryo lethal.||[http://arabidopsis.org/servlets/TairObject?id=136108&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | GBF4||Encodes a basic leucine zipper G-box binding factor that can bind to G-box motifs only as heterodimers with GBF2 or GBF3. A single amino acid change can confer G-box binding as homodimers.||[http://arabidopsis.org/servlets/TairObject?id=28911&type=locus TAIR]|| | + | | GBF4||Encodes a basic leucine zipper G-box binding factor that can bind to G-box motifs only as heterodimers with GBF2 or GBF3. A single amino acid change can confer G-box binding as homodimers.||[http://arabidopsis.org/servlets/TairObject?id=28911&type=locus TAIR]|||| |
|- | |- | ||
| − | | GI||Together with CONSTANTS (CO) and FLOWERING LOCUS T (FT), GIGANTEA promotes flowering under long days in a circadian clock-controlled flowering pathway. GI acts earlier than CO and FT in the pathway by increasing CO and FT mRNA abundance. Located in the nucleus. Regulates several developmental processes, including photoperiod-mediated flowering, phytochrome B signaling, circadian clock, carbohydrate metabolism, and cold stress response. The gene's transcription is controlled by the circadian clock and it is post-transcriptionally regulated by light and dark. Forms a complex with FKF1 on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=136959&type=locus TAIR]||L | + | | GI||Together with CONSTANTS (CO) and FLOWERING LOCUS T (FT), GIGANTEA promotes flowering under long days in a circadian clock-controlled flowering pathway. GI acts earlier than CO and FT in the pathway by increasing CO and FT mRNA abundance. Located in the nucleus. Regulates several developmental processes, including photoperiod-mediated flowering, phytochrome B signaling, circadian clock, carbohydrate metabolism, and cold stress response. The gene's transcription is controlled by the circadian clock and it is post-transcriptionally regulated by light and dark. Forms a complex with FKF1 on the CO promoter to regulate CO expression.||[http://arabidopsis.org/servlets/TairObject?id=136959&type=locus TAIR]||L|| |
|- | |- | ||
| − | | GRF1||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower||[http://arabidopsis.org/servlets/TairObject?id=34438&type=locus TAIR]||FPI | + | | GRF1||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower||[http://arabidopsis.org/servlets/TairObject?id=34438&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF10*||Encodes a 14-3-3 protein. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=136501&type=locus TAIR]||FPI | + | | GRF10*||Encodes a 14-3-3 protein. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=136501&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF11*||Encodes a 14-3-3 protein. Binds H+-ATPase in response to blue light.||[http://arabidopsis.org/servlets/TairObject?id=26877&type=locus TAIR]||FPI | + | | GRF11*||Encodes a 14-3-3 protein. Binds H+-ATPase in response to blue light.||[http://arabidopsis.org/servlets/TairObject?id=26877&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF12*||Encodes a 14-3-3 protein.||[http://arabidopsis.org/servlets/TairObject?id=136716&type=locus TAIR]||FPI | + | | GRF12*||Encodes a 14-3-3 protein.||[http://arabidopsis.org/servlets/TairObject?id=136716&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF2||G-box binding factor GF14 omega encoding a 14-3-3 protein||[http://arabidopsis.org/servlets/TairObject?id=30202&type=locus TAIR]||FPI | + | | GRF2||G-box binding factor GF14 omega encoding a 14-3-3 protein||[http://arabidopsis.org/servlets/TairObject?id=30202&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF3||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=32311&type=locus TAIR]||FPI | + | | GRF3||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=32311&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF4||GF14 protein phi chain member of 14-3-3 protein family. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=137475&type=locus TAIR]||FPI | + | | GRF4||GF14 protein phi chain member of 14-3-3 protein family. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1||[http://arabidopsis.org/servlets/TairObject?id=137475&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF5||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=38052&type=locus TAIR]||FPI | + | | GRF5||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=38052&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF6||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=33231&type=locus TAIR]||FPI | + | | GRF6||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=33231&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF7||Encodes GF14 ν, a 14-3-3 protein isoform (14-3-3ν).||[http://arabidopsis.org/servlets/TairObject?id=36055&type=locus TAIR]||FPI | + | | GRF7||Encodes GF14 ν, a 14-3-3 protein isoform (14-3-3ν).||[http://arabidopsis.org/servlets/TairObject?id=36055&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | GRF8||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=129479&type=locus TAIR]||FPI | + | | GRF8||Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in shoot and flower.||[http://arabidopsis.org/servlets/TairObject?id=129479&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | HAP3A||Encodes a transcription factor from the nuclear factor Y (NF-Y) family, AtNF-YB1. Confers drought tolerance.||[http://arabidopsis.org/servlets/TairObject?id=35360&type=locus TAIR]||FPI | + | | HAP3A||Encodes a transcription factor from the nuclear factor Y (NF-Y) family, AtNF-YB1. Confers drought tolerance.||[http://arabidopsis.org/servlets/TairObject?id=35360&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | HAP3B|| | + | | HAP3B||||||FPI|| |
|- | |- | ||
| − | | HAP5A||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5A) and expressed in vegetative and reproductive tissues.||[http://arabidopsis.org/servlets/TairObject?id=126547&type=locus TAIR]||FPI | + | | HAP5A||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5A) and expressed in vegetative and reproductive tissues.||[http://arabidopsis.org/servlets/TairObject?id=126547&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | HAP5B||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5B) and expressed in vegetative and reproductive tissues||[http://arabidopsis.org/servlets/TairObject?id=27444&type=locus TAIR]||FPI | + | | HAP5B||Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5B) and expressed in vegetative and reproductive tissues||[http://arabidopsis.org/servlets/TairObject?id=27444&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | HAP5C||Heme activated protein (HAP5c)||[http://arabidopsis.org/servlets/TairObject?id=30861&type=locus TAIR]||FPI | + | | HAP5C||Heme activated protein (HAP5c)||[http://arabidopsis.org/servlets/TairObject?id=30861&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | HUA2||Putative transcription factor. Member of the floral homeotic AGAMOUS pathway.Mutations in HUA enhance the phenotype of mild ag-4 allele. Single hua mutants are early flowering and have reduced levels of FLC mRNA. Other MADS box flowering time genes such as FLM and MAF2 also appear to be regulated by HUA2. HUA2 normally activates FLC expression and enhances AG function.||[http://arabidopsis.org/servlets/TairObject?id=135220&type=locus TAIR]||V | + | | HUA2||Putative transcription factor. Member of the floral homeotic AGAMOUS pathway.Mutations in HUA enhance the phenotype of mild ag-4 allele. Single hua mutants are early flowering and have reduced levels of FLC mRNA. Other MADS box flowering time genes such as FLM and MAF2 also appear to be regulated by HUA2. HUA2 normally activates FLC expression and enhances AG function.||[http://arabidopsis.org/servlets/TairObject?id=135220&type=locus TAIR]||V|| |
|- | |- | ||
| − | | LD||Encodes a nuclear localized protein with similarity to transcriptional regulators. Recessive mutants are late flowering. Expression of LFY is reduced in LD mutants.||[http://arabidopsis.org/servlets/TairObject?id=26564&type=locus TAIR]||Au | + | | LD||Encodes a nuclear localized protein with similarity to transcriptional regulators. Recessive mutants are late flowering. Expression of LFY is reduced in LD mutants.||[http://arabidopsis.org/servlets/TairObject?id=26564&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | LDL1*||Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering time loci FLC and FWA. Located in nucleus. Negatively regulates root elongation. Involved in repression of LRP1 via histone deacetylation.||[http://arabidopsis.org/servlets/TairObject?id=29274&type=locus TAIR]||Am, Au | + | | LDL1*||Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering time loci FLC and FWA. Located in nucleus. Negatively regulates root elongation. Involved in repression of LRP1 via histone deacetylation.||[http://arabidopsis.org/servlets/TairObject?id=29274&type=locus TAIR]||Am, Au|| |
|- | |- | ||
| − | | LDL2*||Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering loci FLC and FWA.||[http://arabidopsis.org/servlets/TairObject?id=38624 TAIR]||Am, Au | + | | LDL2*||Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering loci FLC and FWA.||[http://arabidopsis.org/servlets/TairObject?id=38624 TAIR]||Am, Au|| |
|- | |- | ||
| − | | LFY||Encodes transcriptional regulator that promotes the transition to flowering.Involved in floral meristem development. LFY is involved in the regulation of AP3 expression, and appears to bring the F-box protein UFO to the AP3 promoter. Amino acids 46-120 define a protein domain that mediates self-interaction.||[http://arabidopsis.org/servlets/TairObject?id=132616&type=locus TAIR]||FPI, MI | + | | LFY||Encodes transcriptional regulator that promotes the transition to flowering.Involved in floral meristem development. LFY is involved in the regulation of AP3 expression, and appears to bring the F-box protein UFO to the AP3 promoter. Amino acids 46-120 define a protein domain that mediates self-interaction.||[http://arabidopsis.org/servlets/TairObject?id=132616&type=locus TAIR]||FPI, MI||[http://www.uniprot.org/uniprot/Q8L5W6 Grape] |
|- | |- | ||
| − | | LHP1||Regulates the meristem response to light signals and the maintenance of inflorescence meristem identity. Influences developmental processes controlled by APETALA1. TFL2 silences specific genes within euchromatin but not genes positioned in heterochromatin. TFL2 protein localized preferentially to euchromatic regions and not to heterochromatic chromocenters. Involved in euchromatin organization. Required for epigenetic maintenance of the vernalized state.||[http://arabidopsis.org/servlets/TairObject?id=134923&type=locus TAIR]|| | + | | LHP1||Regulates the meristem response to light signals and the maintenance of inflorescence meristem identity. Influences developmental processes controlled by APETALA1. TFL2 silences specific genes within euchromatin but not genes positioned in heterochromatin. TFL2 protein localized preferentially to euchromatic regions and not to heterochromatic chromocenters. Involved in euchromatin organization. Required for epigenetic maintenance of the vernalized state.||[http://arabidopsis.org/servlets/TairObject?id=134923&type=locus TAIR]|||| |
|- | |- | ||
| − | | LHY||LHY encodes a myb-related putative transcription factor involved in circadian rhythm along with another myb transcription factor CCA1.||[http://arabidopsis.org/servlets/TairObject?id=137165&type=locus TAIR]||L | + | | LHY||LHY encodes a myb-related putative transcription factor involved in circadian rhythm along with another myb transcription factor CCA1.||[http://arabidopsis.org/servlets/TairObject?id=137165&type=locus TAIR]||L|| |
|- | |- | ||
| − | | LKP2||Encodes a member of F-box proteins that includes two other proteins in Arabidopsis (ZTL and FKF1). These proteins contain a unique structure containing a PAS domain at their N-terminus, an F-box motif, and 6 kelch repeats at their C-terminus. Overexpression results in arrhythmic phenotypes for a number of circadian clock outputs in both constant light and constant darkness, long hypocotyls under multiple fluences of both red and blue light, and a loss of photoperiodic control of flowering time. Although this the expression of this gene itself is not regulated by circadian clock, it physically interacts with Dof transcription factors that are transcriptionally regulated by circadian rhythm. LKP2 interacts with Di19, CO/COL family proteins.||[http://arabidopsis.org/servlets/TairObject?id=500231567&type=locus TAIR]||L | + | | LKP2||Encodes a member of F-box proteins that includes two other proteins in Arabidopsis (ZTL and FKF1). These proteins contain a unique structure containing a PAS domain at their N-terminus, an F-box motif, and 6 kelch repeats at their C-terminus. Overexpression results in arrhythmic phenotypes for a number of circadian clock outputs in both constant light and constant darkness, long hypocotyls under multiple fluences of both red and blue light, and a loss of photoperiodic control of flowering time. Although this the expression of this gene itself is not regulated by circadian clock, it physically interacts with Dof transcription factors that are transcriptionally regulated by circadian rhythm. LKP2 interacts with Di19, CO/COL family proteins.||[http://arabidopsis.org/servlets/TairObject?id=500231567&type=locus TAIR]||L|| |
|- | |- | ||
| − | | LUX||Encodes a myb family transcription factor with a single Myb DNA-binding domain (type SHAQKYF) that is unique to plants and is essential for circadian rhythms, specifically for transcriptional regulation within the circadian clock. LUX is required for normal rhythmic expression of multiple clock outputs in both constant light and darkness. It is coregulated with TOC1 and seems to be repressed by CCA1 and LHY by direct binding of these proteins to the evening element in the LUX promoter.||[http://arabidopsis.org/servlets/TairObject?id=35747&type=locus TAIR]||L | + | | LUX||Encodes a myb family transcription factor with a single Myb DNA-binding domain (type SHAQKYF) that is unique to plants and is essential for circadian rhythms, specifically for transcriptional regulation within the circadian clock. LUX is required for normal rhythmic expression of multiple clock outputs in both constant light and darkness. It is coregulated with TOC1 and seems to be repressed by CCA1 and LHY by direct binding of these proteins to the evening element in the LUX promoter.||[http://arabidopsis.org/servlets/TairObject?id=35747&type=locus TAIR]||L|| |
|- | |- | ||
| − | | MAF1||MADS domain protein - flowering regulator that is closely related to FLC. Deletion of this locus in Nd ecotype is correlated with earlier flowering in short days suggesting function as a negative regulator of flowering.||[http://arabidopsis.org/servlets/TairObject?id=29106&type=locus TAIR]||Am, FPI, V | + | | MAF1||MADS domain protein - flowering regulator that is closely related to FLC. Deletion of this locus in Nd ecotype is correlated with earlier flowering in short days suggesting function as a negative regulator of flowering.||[http://arabidopsis.org/servlets/TairObject?id=29106&type=locus TAIR]||Am, FPI, V|| |
|- | |- | ||
| − | | MAF3||MADS domain protein - flowering regulator that is closely related to FLC||[http://arabidopsis.org/servlets/TairObject?id=130633&type=locus TAIR]||Am, FPI, V | + | | MAF3||MADS domain protein - flowering regulator that is closely related to FLC||[http://arabidopsis.org/servlets/TairObject?id=130633&type=locus TAIR]||Am, FPI, V|| |
|- | |- | ||
| − | | MAF4||Encodes MADS-box containing FLC paralog. Five splice variants have been identified but not characterized with respect to expression patterns and/or differing function. Overexpression of the gene in the Landsberg ecotype leads to a delay in flowering, transcript levels of MAF4 are reduced after a 6 week vernalization.||[http://arabidopsis.org/servlets/TairObject?id=130634&type=locus TAIR]||Am, FPI, V | + | | MAF4||Encodes MADS-box containing FLC paralog. Five splice variants have been identified but not characterized with respect to expression patterns and/or differing function. Overexpression of the gene in the Landsberg ecotype leads to a delay in flowering, transcript levels of MAF4 are reduced after a 6 week vernalization.||[http://arabidopsis.org/servlets/TairObject?id=130634&type=locus TAIR]||Am, FPI, V|| |
|- | |- | ||
| − | | MAF5||Is upregulated during vernalization and regulates flowering time. Encodes MADS-domain protein. Two variants encoding proteins of 198 and 184 amino acids have been reported.||[http://arabidopsis.org/servlets/TairObject?id=130635&type=locus TAIR]||Am, FPI, V | + | | MAF5||Is upregulated during vernalization and regulates flowering time. Encodes MADS-domain protein. Two variants encoding proteins of 198 and 184 amino acids have been reported.||[http://arabidopsis.org/servlets/TairObject?id=130635&type=locus TAIR]||Am, FPI, V|| |
|- | |- | ||
| − | | MEA*||Encodes a putative transcription factor MEDEA (MEA) that negatively regulates seed development in the absence of fertilization. Mutations in this locus result in embryo lethality. MEA is a Polycomb group gene that is imprinted in the endosperm. The maternal allele is expressed and the paternal allele is silent. MEA is controlled by DEMETER (DME), a DNA glycosylase required to activate MEA expression, and METHYLTRANSFERASE I (MET1), which maintains CG methylation at the MEA locus. MEA is involved in the negative regulation of its own imprinted gene expression; the effect is not only allele-specific but also dynamically regulated during seed development. In the ovule, the MEA transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization||[http://arabidopsis.org/servlets/TairObject?id=136450&type=locus TAIR]||Au, V | + | | MEA*||Encodes a putative transcription factor MEDEA (MEA) that negatively regulates seed development in the absence of fertilization. Mutations in this locus result in embryo lethality. MEA is a Polycomb group gene that is imprinted in the endosperm. The maternal allele is expressed and the paternal allele is silent. MEA is controlled by DEMETER (DME), a DNA glycosylase required to activate MEA expression, and METHYLTRANSFERASE I (MET1), which maintains CG methylation at the MEA locus. MEA is involved in the negative regulation of its own imprinted gene expression; the effect is not only allele-specific but also dynamically regulated during seed development. In the ovule, the MEA transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization||[http://arabidopsis.org/servlets/TairObject?id=136450&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | MSI1||Encodes a WD-40 repeat containing protein that functions in chromatin assembly as part of the CAF1 and FIE complex. Mutants exhibit parthenogenetic development that includes proliferation of unfertilized endosperm and embryos. In heterozygous plants 50% of embryos abort. Of the aborted embryos the early aborted class are homozygous and the later aborting lass are heterozygotes in which the defective allele is maternally transmitted. Other phenotypes include defects in ovule morphogenesis and organ initiation,as well as increased levels of heterochromatic DNA. MSI1 is needed for the transition to flowering. In Arabidopsis, the three CAF-1 subunits are encoded by FAS1, FAS2 and, most likely, MSI1, respectively. Mutations in FAS1 or FAS2 lead to increased frequency of homologous recombination and T-DNA integration in Arabidopsis. In the ovule, the MSI1 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization. MSI is biallelically expressed, the paternall allele is expressed in the endosperm and embryo and is not imprinted. MSI1 forms a complex with RBR1 that is required for activation of the imprinted genes FIS2 and FWA. This activation is mediated by MSI1/RBR1 mediated repression of MET1.||[http://arabidopsis.org/servlets/TairObject?id=132921&type=locus TAIR]||Au, V | + | | MSI1||Encodes a WD-40 repeat containing protein that functions in chromatin assembly as part of the CAF1 and FIE complex. Mutants exhibit parthenogenetic development that includes proliferation of unfertilized endosperm and embryos. In heterozygous plants 50% of embryos abort. Of the aborted embryos the early aborted class are homozygous and the later aborting lass are heterozygotes in which the defective allele is maternally transmitted. Other phenotypes include defects in ovule morphogenesis and organ initiation,as well as increased levels of heterochromatic DNA. MSI1 is needed for the transition to flowering. In Arabidopsis, the three CAF-1 subunits are encoded by FAS1, FAS2 and, most likely, MSI1, respectively. Mutations in FAS1 or FAS2 lead to increased frequency of homologous recombination and T-DNA integration in Arabidopsis. In the ovule, the MSI1 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization. MSI is biallelically expressed, the paternall allele is expressed in the endosperm and embryo and is not imprinted. MSI1 forms a complex with RBR1 that is required for activation of the imprinted genes FIS2 and FWA. This activation is mediated by MSI1/RBR1 mediated repression of MET1.||[http://arabidopsis.org/servlets/TairObject?id=132921&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | MSI2*||Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA||[http://www.uniprot.org/uniprot/O22468 UniProtKB]||Au, V | + | | MSI2*||Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA||[http://www.uniprot.org/uniprot/O22468 UniProtKB]||Au, V|| |
|- | |- | ||
| − | | MSI3*||Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA||[http://www.uniprot.org/uniprot/O22469 UniProtKB]||Au, V | + | | MSI3*||Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA||[http://www.uniprot.org/uniprot/O22469 UniProtKB]||Au, V|| |
|- | |- | ||
| − | | NF-YB10||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=37281&type=locus TAIR]||FPI | + | | NF-YB10||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=37281&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NF-YB3||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=128758&type=locus TAIR]||FPI | + | | NF-YB3||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=128758&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NF-YB7||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=33626&type=locus TAIR]||FPI | + | | NF-YB7||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=33626&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NF-YB8||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=34862&type=locus TAIR]||FPI | + | | NF-YB8||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=34862&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NF-YC3||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=27340&type=locus TAIR]||FPI | + | | NF-YC3||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=27340&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NF-YC4||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=133713&type=locus TAIR]||FPI | + | | NF-YC4||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=133713&type=locus TAIR]||FPI|| |
|- | |- | ||
| − | | NFC5*||Cell cycle-related repressor genes encoding WD-repeat proteins.||[http://arabidopsis.org/servlets/TairObject?id=129407&type=locus TAIR]||Am, Au | + | | NFC5*||Cell cycle-related repressor genes encoding WD-repeat proteins.||[http://arabidopsis.org/servlets/TairObject?id=129407&type=locus TAIR]||Am, Au|| |
|- | |- | ||
| − | | PAF1||Encodes a protein with extensive homology to the largest subunit of the multicatalytic proteinase complex (proteasome). Negatively regulates thiol biosynthesis and arsenic tolerance.||[http://arabidopsis.org/servlets/TairObject?id=133505&type=locus TAIR]||V | + | | PAF1||Encodes a protein with extensive homology to the largest subunit of the multicatalytic proteinase complex (proteasome). Negatively regulates thiol biosynthesis and arsenic tolerance.||[http://arabidopsis.org/servlets/TairObject?id=133505&type=locus TAIR]||V|| |
|- | |- | ||
| − | | PAF2||Encodes 20S proteasome subunit PAF2 (PAF2).||[http://arabidopsis.org/servlets/TairObject?id=30983&type=locus TAIR]||V | + | | PAF2||Encodes 20S proteasome subunit PAF2 (PAF2).||[http://arabidopsis.org/servlets/TairObject?id=30983&type=locus TAIR]||V|| |
|- | |- | ||
| − | | PEP||Encodes a novel Arabidopsis gene encoding a polypeptide with K-homology (KH) RNA-binding modules, which acts on vegetative growth and pistil development. Genetic studies suggest that PEP interacts with element(s) of the CLAVATA signaling pathway.||[http://arabidopsis.org/servlets/TairObject?id=127403&type=locus TAIR]||V | + | | PEP||Encodes a novel Arabidopsis gene encoding a polypeptide with K-homology (KH) RNA-binding modules, which acts on vegetative growth and pistil development. Genetic studies suggest that PEP interacts with element(s) of the CLAVATA signaling pathway.||[http://arabidopsis.org/servlets/TairObject?id=127403&type=locus TAIR]||V|| |
|- | |- | ||
| − | | PFT1||Encodes a nuclear protein that acts in a phyB pathway (but downstream of phyB) and induces flowering in response to suboptimal light conditions. PFT1 promotes flowering in CO dependent and independent pathways and integrates several environmental stimuli, such as light quality and JA-dependent defenses. Mutants are hypo-responsive to far-red and hyper-responsive to red light and flower late under long day conditions. Also shown to be a Mediator subunit regulating jasmonate-dependent defense.||[http://arabidopsis.org/servlets/TairObject?id=30064&type=locus TAIR]|| | + | | PFT1||Encodes a nuclear protein that acts in a phyB pathway (but downstream of phyB) and induces flowering in response to suboptimal light conditions. PFT1 promotes flowering in CO dependent and independent pathways and integrates several environmental stimuli, such as light quality and JA-dependent defenses. Mutants are hypo-responsive to far-red and hyper-responsive to red light and flower late under long day conditions. Also shown to be a Mediator subunit regulating jasmonate-dependent defense.||[http://arabidopsis.org/servlets/TairObject?id=30064&type=locus TAIR]|||| |
|- | |- | ||
| − | | PHYA||Light-labile cytoplasmic red/far-red light photoreceptor involved in the regulation of photomorphogenesis. It exists in two inter-convertible forms: Pr and Pfr (active) and functions as a dimer.The N terminus carries a single tetrapyrrole chromophore, and the C terminus is involved in dimerization. It is the sole photoreceptor mediating the FR high irradiance response (HIR). Major regulator in red-light induction of phototropic enhancement. Involved in the regulation of de-etiolation. Involved in gravitropism and phototropism. Requires FHY1 for nuclear accumulation.||[http://arabidopsis.org/servlets/TairObject?id=27545&type=locus TAIR]||L | + | | PHYA||Light-labile cytoplasmic red/far-red light photoreceptor involved in the regulation of photomorphogenesis. It exists in two inter-convertible forms: Pr and Pfr (active) and functions as a dimer.The N terminus carries a single tetrapyrrole chromophore, and the C terminus is involved in dimerization. It is the sole photoreceptor mediating the FR high irradiance response (HIR). Major regulator in red-light induction of phototropic enhancement. Involved in the regulation of de-etiolation. Involved in gravitropism and phototropism. Requires FHY1 for nuclear accumulation.||[http://arabidopsis.org/servlets/TairObject?id=27545&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/A5B9C2 Grape] |
|- | |- | ||
| − | | PHYB||Red/far-red photoreceptor involved in the regulation of de-etiolation. Exists in two inter-convertible forms: Pr and Pfr (active). Involved in the light-promotion of seed germination and in the shade avoidance response. Promotes seedling etiolation in both the presence and absence of phytochrome A. Overexpression results in etiolation under far-red light. Accumulates in the nucleus after exposure to far red light.||[http://arabidopsis.org/servlets/TairObject?id=26548&type=locus TAIR]||L | + | | PHYB||Red/far-red photoreceptor involved in the regulation of de-etiolation. Exists in two inter-convertible forms: Pr and Pfr (active). Involved in the light-promotion of seed germination and in the shade avoidance response. Promotes seedling etiolation in both the presence and absence of phytochrome A. Overexpression results in etiolation under far-red light. Accumulates in the nucleus after exposure to far red light.||[http://arabidopsis.org/servlets/TairObject?id=26548&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/B9U4G3 Grape] |
|- | |- | ||
| − | | PHYC||Encodes the apoprotein of phytochrome;one of a family of photoreceptors that modulate plant growth and development.||[http://arabidopsis.org/servlets/TairObject?id=133466&type=locus TAIR]||L | + | | PHYC||Encodes the apoprotein of phytochrome;one of a family of photoreceptors that modulate plant growth and development.||[http://arabidopsis.org/servlets/TairObject?id=133466&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/B9U4G4 Grape] |
|- | |- | ||
| − | | PHYD||Encodes a phytochrome photoreceptor with a function similar to that of phyB that absorbs the red/far-red part of the light spectrum and is involved in light responses. It cannot compensate for phyB loss in Arabidopsis but can substitute for tobacco phyB in vivo.||[http://arabidopsis.org/servlets/TairObject?id=26567&type=locus TAIR]||L | + | | PHYD||Encodes a phytochrome photoreceptor with a function similar to that of phyB that absorbs the red/far-red part of the light spectrum and is involved in light responses. It cannot compensate for phyB loss in Arabidopsis but can substitute for tobacco phyB in vivo.||[http://arabidopsis.org/servlets/TairObject?id=26567&type=locus TAIR]||L|| |
|- | |- | ||
| − | | PHYE||Histidine kinase||[http://arabidopsis.org/servlets/TairObject?id=26568&type=locus TAIR]||L | + | | PHYE||Histidine kinase||[http://arabidopsis.org/servlets/TairObject?id=26568&type=locus TAIR]||L||[http://www.uniprot.org/uniprot/B9U4G5 Grape] |
|- | |- | ||
| − | | PI||Floral homeotic gene encoding a MADS domain transcription factor. Required for the specification of petal and stamen identities.||[http://arabidopsis.org/servlets/TairObject?id=131273&type=locus TAIR]|| | + | | PI||Floral homeotic gene encoding a MADS domain transcription factor. Required for the specification of petal and stamen identities.||[http://arabidopsis.org/servlets/TairObject?id=131273&type=locus TAIR]||||[http://www.uniprot.org/uniprot/J9SQF2 Grape] |
|- | |- | ||
| − | | PIE1||Encodes a protein similar to ATP-dependent, chromatin-remodeling proteins of the ISWI and SWI2/SNF2 family. Genetic analyses suggest that this gene is involved in multiple flowering pathways. Mutations in PIE1 results in suppression of FLC-mediated delay of flowering and causes early flowering in noninductive photoperiods independently of FLC. PIE1 is required for expression of FLC in the shoot apex but not in the root.Along with ARP6 forms a complex to deposit modified histone H2A.Z at several loci within the genome. This modification alters the expression of the target genes (i.e. FLC, MAF4, MAF6).||[http://arabidopsis.org/servlets/TairObject?id=37964&type=locus TAIR]||V | + | | PIE1||Encodes a protein similar to ATP-dependent, chromatin-remodeling proteins of the ISWI and SWI2/SNF2 family. Genetic analyses suggest that this gene is involved in multiple flowering pathways. Mutations in PIE1 results in suppression of FLC-mediated delay of flowering and causes early flowering in noninductive photoperiods independently of FLC. PIE1 is required for expression of FLC in the shoot apex but not in the root.Along with ARP6 forms a complex to deposit modified histone H2A.Z at several loci within the genome. This modification alters the expression of the target genes (i.e. FLC, MAF4, MAF6).||[http://arabidopsis.org/servlets/TairObject?id=37964&type=locus TAIR]||V|| |
|- | |- | ||
| − | | PIF3||Transcription factor interacting with photoreceptors phyA and phyB. Forms a ternary complex in vitro with G-box element of the promoters of LHY, CCA1. Acts as a negative regulator of phyB signalling. It degrades rapidly after irradiation of dark grown seedlings in a process controlled by phytochromes. Does not play a significant role in controlling light input and function of the circadian clockwork. Binds to G- and E-boxes, but not to other ACEs. Binds to anthocyanin biosynthetic genes in a light- and HY5-independent fashion. PIF3 function as a transcriptional activator can be functionally and mechanistically separated from its role in repression of PhyB mediated processes.||[http://arabidopsis.org/servlets/TairObject?id=27554&type=locus TAIR]|| | + | | PIF3||Transcription factor interacting with photoreceptors phyA and phyB. Forms a ternary complex in vitro with G-box element of the promoters of LHY, CCA1. Acts as a negative regulator of phyB signalling. It degrades rapidly after irradiation of dark grown seedlings in a process controlled by phytochromes. Does not play a significant role in controlling light input and function of the circadian clockwork. Binds to G- and E-boxes, but not to other ACEs. Binds to anthocyanin biosynthetic genes in a light- and HY5-independent fashion. PIF3 function as a transcriptional activator can be functionally and mechanistically separated from its role in repression of PhyB mediated processes.||[http://arabidopsis.org/servlets/TairObject?id=27554&type=locus TAIR]|||| |
|- | |- | ||
| − | | PRR7||PRR7 and PRR9 are partially redundant essential components of a temperature-sensitive circadian system. CCA1 and LHY had a positive effect on PRR7 expression levels. Acts as transcriptional repressor of CCA1 and LHY.||[http://arabidopsis.org/servlets/TairObject?id=131538&type=locus TAIR]||L | + | | PRR7||PRR7 and PRR9 are partially redundant essential components of a temperature-sensitive circadian system. CCA1 and LHY had a positive effect on PRR7 expression levels. Acts as transcriptional repressor of CCA1 and LHY.||[http://arabidopsis.org/servlets/TairObject?id=131538&type=locus TAIR]||L|| |
|- | |- | ||
| − | | RAV1||Encodes an AP2/B3 domain transcription factor which is upregulated in response to low temperature. It contains a B3 DNA binding domain. It has circadian regulation and may function as a negative growth regulator.||[http://arabidopsis.org/servlets/TairObject?id=137721&type=locus TAIR]||L | + | | RAV1||Encodes an AP2/B3 domain transcription factor which is upregulated in response to low temperature. It contains a B3 DNA binding domain. It has circadian regulation and may function as a negative growth regulator.||[http://arabidopsis.org/servlets/TairObject?id=137721&type=locus TAIR]||L|| |
|- | |- | ||
| − | | REF6||Relative of Early Flowering 6 (REF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a positive regulator of flowering in an FLC-dependent pathway. REF6 mutants have hyperacetylation of histone H4 at the FLC locus. REF6 interacts with BES1 in a Y2H assay and in vitro. REF6 may play a role in brassinoteroid signaling by affecting histone methylation in the promoters of BR-responsive genes. It is most closely related to the JHDM3 subfamily of JmjN/C proteins.||[http://arabidopsis.org/servlets/TairObject?id=500230554&type=locus TAIR]|| | + | | REF6||Relative of Early Flowering 6 (REF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a positive regulator of flowering in an FLC-dependent pathway. REF6 mutants have hyperacetylation of histone H4 at the FLC locus. REF6 interacts with BES1 in a Y2H assay and in vitro. REF6 may play a role in brassinoteroid signaling by affecting histone methylation in the promoters of BR-responsive genes. It is most closely related to the JHDM3 subfamily of JmjN/C proteins.||[http://arabidopsis.org/servlets/TairObject?id=500230554&type=locus TAIR]|||| |
|- | |- | ||
| − | | RFI2||Zinc ion binding||[http://arabidopsis.org/servlets/TairObject?id=32070&type=locus TAIR]||L | + | | RFI2||Zinc ion binding||[http://arabidopsis.org/servlets/TairObject?id=32070&type=locus TAIR]||L|| |
|- | |- | ||
| − | | SEC*||Has O-linked N-acetyl glucosamine transferase activity. Similar to Arabidopsis SPY gene.||[http://arabidopsis.org/servlets/TairObject?id=40600&type=locus TAIR]|| | + | | SEC*||Has O-linked N-acetyl glucosamine transferase activity. Similar to Arabidopsis SPY gene.||[http://arabidopsis.org/servlets/TairObject?id=40600&type=locus TAIR]|||| |
|- | |- | ||
| − | | SEP1||Encodes a stress enhanced protein that localizes to the thylakoid membrane and whose mRNA is upregulated in response to high light intensity. It may be involved in chlorophyll binding.||[http://arabidopsis.org/servlets/TairObject?id=127910&type=locus TAIR]|| | + | | SEP1||Encodes a stress enhanced protein that localizes to the thylakoid membrane and whose mRNA is upregulated in response to high light intensity. It may be involved in chlorophyll binding.||[http://arabidopsis.org/servlets/TairObject?id=127910&type=locus TAIR]|||| |
|- | |- | ||
| − | | SEP2||Stress enhanced protein 2 (SEP2) chlorophyll a/b-binding protein||[http://arabidopsis.org/servlets/TairObject?id=33372&type=locus TAIR]|| | + | | SEP2||Stress enhanced protein 2 (SEP2) chlorophyll a/b-binding protein||[http://arabidopsis.org/servlets/TairObject?id=33372&type=locus TAIR]|||| |
|- | |- | ||
| − | | SEP3||Member of the MADs box transcription factor family. SEP3 is redundant with SEP1 and 2. Flowers of SEP1/2/3 triple mutants show a conversion of petals and stamens to sepals.SEP3 forms heterotetrameric complexes with other MADS box family members and binds to the CArG box motif.||[http://arabidopsis.org/servlets/TairObject?id=30252&type=locus TAIR]|| | + | | SEP3||Member of the MADs box transcription factor family. SEP3 is redundant with SEP1 and 2. Flowers of SEP1/2/3 triple mutants show a conversion of petals and stamens to sepals.SEP3 forms heterotetrameric complexes with other MADS box family members and binds to the CArG box motif.||[http://arabidopsis.org/servlets/TairObject?id=30252&type=locus TAIR]|||| |
|- | |- | ||
| − | | SEP4||This gene belongs to the family of SEP genes. It is involved in the development of sepals, petals, stamens and carpels. Additionally, it plays a central role in the determination of flower meristem and organ identity.||[http://arabidopsis.org/servlets/TairObject?id=32207&type=locus TAIR]|| | + | | SEP4||This gene belongs to the family of SEP genes. It is involved in the development of sepals, petals, stamens and carpels. Additionally, it plays a central role in the determination of flower meristem and organ identity.||[http://arabidopsis.org/servlets/TairObject?id=32207&type=locus TAIR]|||| |
|- | |- | ||
| − | | SHP1*||One of two genes (SHP1 and SHP2) that are required for fruit dehiscence. The two genes control dehiscence zone differentiation and promote the lignification of adjacent cells.||[http://arabidopsis.org/servlets/TairObject?id=39912&type=locus TAIR]|| | + | | SHP1*||One of two genes (SHP1 and SHP2) that are required for fruit dehiscence. The two genes control dehiscence zone differentiation and promote the lignification of adjacent cells.||[http://arabidopsis.org/servlets/TairObject?id=39912&type=locus TAIR]|||| |
|- | |- | ||
| − | | SHP2*||AGAMOUS [AG]-like MADS box protein (AGL5) involved in fruit development (valve margin and dehiscence zone differentiation). A putative direct target of AG. SHP2 has been shown to be a downstream gene of the complex formed by AG and SEP proteins (SEP4 alone does not form a functional complex with AG).||[http://arabidopsis.org/servlets/TairObject?id=33362&type=locus TAIR]|| | + | | SHP2*||AGAMOUS [AG]-like MADS box protein (AGL5) involved in fruit development (valve margin and dehiscence zone differentiation). A putative direct target of AG. SHP2 has been shown to be a downstream gene of the complex formed by AG and SEP proteins (SEP4 alone does not form a functional complex with AG).||[http://arabidopsis.org/servlets/TairObject?id=33362&type=locus TAIR]|||| |
|- | |- | ||
| − | | SMZ||Encodes a AP2 domain transcription factor that can repress flowering. SMZ and its paralogous gene, SNARCHZAPFEN (SNZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering.||[http://arabidopsis.org/servlets/TairObject?id=37065&type=locus TAIR]|| | + | | SMZ||Encodes a AP2 domain transcription factor that can repress flowering. SMZ and its paralogous gene, SNARCHZAPFEN (SNZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering.||[http://arabidopsis.org/servlets/TairObject?id=37065&type=locus TAIR]|||| |
|- | |- | ||
| − | | SNZ||Encodes a AP2 domain transcription factor that can repress flowering. SNZ and its paralogous gene, SCHLAFMUTZE (SMZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering.||[http://arabidopsis.org/servlets/TairObject?id=33943&type=locus TAIR]|| | + | | SNZ||Encodes a AP2 domain transcription factor that can repress flowering. SNZ and its paralogous gene, SCHLAFMUTZE (SMZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering.||[http://arabidopsis.org/servlets/TairObject?id=33943&type=locus TAIR]|||| |
|- | |- | ||
| − | | SOC1||Transcription activator active in flowering time control. May integrate signals from the photoperiod, vernalization and autonomous floral induction pathways. Can modulate class B and C homeotic genes expression. When associated with AGL24, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development.||[http://www.uniprot.org/uniprot/O64645 UniProtKB]||FPI | + | | SOC1||Transcription activator active in flowering time control. May integrate signals from the photoperiod, vernalization and autonomous floral induction pathways. Can modulate class B and C homeotic genes expression. When associated with AGL24, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development.||[http://www.uniprot.org/uniprot/O64645 UniProtKB]||FPI||[http://www.uniprot.org/uniprot/D1MDP7 Grape] |
|- | |- | ||
| − | | SPA1||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA1 is a PHYA signaling intermediate, putative regulator of PHYA signaling pathway. Light responsive repressor of photomorphogenesis. Involved in regulating circadian rhythms and flowering time in plants. Under constant light, the abundance of SPA1 protein exhibited circadian regulation, whereas under constant darkness, SPA1 protein levels remained unchanged. In addition, the spa1-3 mutation slightly shortened circadian period of CCA1, TOC1/PRR1 and SPA1 transcript accumulation under constant light.||[http://arabidopsis.org/servlets/TairObject?id=31336&type=locus TAIR]||L | + | | SPA1||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA1 is a PHYA signaling intermediate, putative regulator of PHYA signaling pathway. Light responsive repressor of photomorphogenesis. Involved in regulating circadian rhythms and flowering time in plants. Under constant light, the abundance of SPA1 protein exhibited circadian regulation, whereas under constant darkness, SPA1 protein levels remained unchanged. In addition, the spa1-3 mutation slightly shortened circadian period of CCA1, TOC1/PRR1 and SPA1 transcript accumulation under constant light.||[http://arabidopsis.org/servlets/TairObject?id=31336&type=locus TAIR]||L|| |
|- | |- | ||
| − | | SPA2||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA2 primarily regulates seedling development in darkness and has little function in light-grown seedlings or adult plants.||[http://arabidopsis.org/servlets/TairObject?id=129633&type=locus TAIR]||L | + | | SPA2||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA2 primarily regulates seedling development in darkness and has little function in light-grown seedlings or adult plants.||[http://arabidopsis.org/servlets/TairObject?id=129633&type=locus TAIR]||L|| |
|- | |- | ||
| − | | SPA3||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA3 (and SPA4) predominantly regulates elongation growth in adult plants.||[http://arabidopsis.org/servlets/TairObject?id=500230681&type=locus TAIR]||L | + | | SPA3||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA3 (and SPA4) predominantly regulates elongation growth in adult plants.||[http://arabidopsis.org/servlets/TairObject?id=500230681&type=locus TAIR]||L|| |
|- | |- | ||
| − | | SPA4||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA4 (and SPA3) predominantly regulates elongation growth in adult plants.||[http://arabidopsis.org/servlets/TairObject?id=31031&type=locus TAIR]||L | + | | SPA4||Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA4 (and SPA3) predominantly regulates elongation growth in adult plants.||[http://arabidopsis.org/servlets/TairObject?id=31031&type=locus TAIR]||L|| |
|- | |- | ||
| − | | SPL3||Encodes a member of the SPL (squamosa-promoter binding protein-like)gene family, a novel gene family encoding DNA binding proteins and putative transcription factors. Contains the SBP-box, which encodes the SBP-domain, required and sufficient for interaction with DNA. It binds DNA, may directly regulate AP1, and is involved in regulation of flowering and vegetative phase change. Its temporal expression is regulated by the microRNA miR156. The target site for the microRNA is in the 3'UTR.||[http://arabidopsis.org/servlets/TairObject?id=34194&type=locus TAIR]||MI | + | | SPL3||Encodes a member of the SPL (squamosa-promoter binding protein-like)gene family, a novel gene family encoding DNA binding proteins and putative transcription factors. Contains the SBP-box, which encodes the SBP-domain, required and sufficient for interaction with DNA. It binds DNA, may directly regulate AP1, and is involved in regulation of flowering and vegetative phase change. Its temporal expression is regulated by the microRNA miR156. The target site for the microRNA is in the 3'UTR.||[http://arabidopsis.org/servlets/TairObject?id=34194&type=locus TAIR]||MI|| |
|- | |- | ||
| − | | SPY||Encodes a N-acetyl glucosamine transferase that may glycosylate other molecules involved in GA signaling. Contains a tetratricopeptide repeat region, and a novel carboxy-terminal region. SPY acts as both a repressor of GA responses and as a positive regulation of cytokinin signalling. SPY may be involved in reducing ROS accumulation in response to stress.||[http://arabidopsis.org/servlets/TairObject?id=36698&type=locus TAIR]|| | + | | SPY||Encodes a N-acetyl glucosamine transferase that may glycosylate other molecules involved in GA signaling. Contains a tetratricopeptide repeat region, and a novel carboxy-terminal region. SPY acts as both a repressor of GA responses and as a positive regulation of cytokinin signalling. SPY may be involved in reducing ROS accumulation in response to stress.||[http://arabidopsis.org/servlets/TairObject?id=36698&type=locus TAIR]|||| |
|- | |- | ||
| − | | STK*||Encodes a MADS box transcription factor expressed in the carpel and ovules. Plays a maternal role in fertilization and seed development.||[http://arabidopsis.org/servlets/TairObject?id=130184&type=locus TAIR]|| | + | | STK*||Encodes a MADS box transcription factor expressed in the carpel and ovules. Plays a maternal role in fertilization and seed development.||[http://arabidopsis.org/servlets/TairObject?id=130184&type=locus TAIR]|||| |
|- | |- | ||
| − | | STM||Class I knotted-like homeodomain protein that is required for shoot apical meristem (SAM) formation during embryogenesis and for SAM function throughout the lifetime of the plant. Functions by preventing incorporation of cells in the meristem center into differentiating organ primordia.||[http://arabidopsis.org/servlets/TairObject?id=29436&type=locus TAIR]|| | + | | STM||Class I knotted-like homeodomain protein that is required for shoot apical meristem (SAM) formation during embryogenesis and for SAM function throughout the lifetime of the plant. Functions by preventing incorporation of cells in the meristem center into differentiating organ primordia.||[http://arabidopsis.org/servlets/TairObject?id=29436&type=locus TAIR]|||| |
|- | |- | ||
| − | | SUF4||Encodes SUF4 (SUPPRESSOR of FRI 4), a putative zinc-finger-containing transcription factor that is required for delayed flowering in winter-annual Arabidopsis. suf4 mutations strongly suppress the late-flowering phenotype of FRI (FRIGIDA) mutants. suf4 mutants also show reduced H3K4 trimethylation at FLC (FLOWERING LOCUS C), a floral inhibitor. SUF4 may act to specifically recruit a putative histone H3 methyltransferase EFS (EARLY FLOWERING IN SHORT DAYS) and the PAF1-like complex to the FLC locus.||[http://arabidopsis.org/servlets/TairObject?id=28096&type=locus TAIR]||V | + | | SUF4||Encodes SUF4 (SUPPRESSOR of FRI 4), a putative zinc-finger-containing transcription factor that is required for delayed flowering in winter-annual Arabidopsis. suf4 mutations strongly suppress the late-flowering phenotype of FRI (FRIGIDA) mutants. suf4 mutants also show reduced H3K4 trimethylation at FLC (FLOWERING LOCUS C), a floral inhibitor. SUF4 may act to specifically recruit a putative histone H3 methyltransferase EFS (EARLY FLOWERING IN SHORT DAYS) and the PAF1-like complex to the FLC locus.||[http://arabidopsis.org/servlets/TairObject?id=28096&type=locus TAIR]||V|| |
|- | |- | ||
| − | | SVP||Encodes a nuclear protein that acts as a floral repressor and that functions within the thermosensory pathway. SVP represses FT expression via direct binding to the vCArG III motif in the FT promoter.||[http://arabidopsis.org/servlets/TairObject?id=31694&type=locus TAIR]||Am, V | + | | SVP||Encodes a nuclear protein that acts as a floral repressor and that functions within the thermosensory pathway. SVP represses FT expression via direct binding to the vCArG III motif in the FT promoter.||[http://arabidopsis.org/servlets/TairObject?id=31694&type=locus TAIR]||Am, V||[http://www.uniprot.org/uniprot/H9CTT8 Grape] |
|- | |- | ||
| − | | SWN||Encodes a polycomb group protein. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and CURLY LEAF (CLF). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. Performs a partially redundant role to MEA in controlling seed initiation by helping to suppress central cell nucleus endosperm proliferation within the FG.||[http://arabidopsis.org/servlets/TairObject?id=129104&type=locus TAIR]||Au, V | + | | SWN||Encodes a polycomb group protein. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and CURLY LEAF (CLF). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. Performs a partially redundant role to MEA in controlling seed initiation by helping to suppress central cell nucleus endosperm proliferation within the FG.||[http://arabidopsis.org/servlets/TairObject?id=129104&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | TEM1||Encodes a member of the RAV transcription factor family that contains AP2 and B3 binding domains. Involved in the regulation of flowering under long days. Loss of function results in early flowering. Overexpression causes late flowering and repression of expression of FT. Novel transcriptional regulator involved in ethylene signaling. Promoter bound by EIN3. EDF1 in turn, binds to promoter elements in ethylene responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=30061&type=locus TAIR]||L | + | | TEM1||Encodes a member of the RAV transcription factor family that contains AP2 and B3 binding domains. Involved in the regulation of flowering under long days. Loss of function results in early flowering. Overexpression causes late flowering and repression of expression of FT. Novel transcriptional regulator involved in ethylene signaling. Promoter bound by EIN3. EDF1 in turn, binds to promoter elements in ethylene responsive genes.||[http://arabidopsis.org/servlets/TairObject?id=30061&type=locus TAIR]||L|| |
|- | |- | ||
| − | | TEM2||Rav2 is part of a complex that has been named `regulator of the (H+)-ATPase of the vacuolar and endosomal membranes' (RAVE)||[http://arabidopsis.org/servlets/TairObject?id=27570&type=locus TAIR]||L | + | | TEM2||Rav2 is part of a complex that has been named `regulator of the (H+)-ATPase of the vacuolar and endosomal membranes' (RAVE)||[http://arabidopsis.org/servlets/TairObject?id=27570&type=locus TAIR]||L|| |
|- | |- | ||
| − | | TFL1||Controls inflorescence meristem identity. Involved in the floral initiation process. Ortholog of the Antirrhinum gene CENTRORADIALIS (CEN). Involved in protein trafficking to the protein storage vacuole.||[http://arabidopsis.org/servlets/TairObject?id=131459&type=locus TAIR]||Am, FPI | + | | TFL1||Controls inflorescence meristem identity. Involved in the floral initiation process. Ortholog of the Antirrhinum gene CENTRORADIALIS (CEN). Involved in protein trafficking to the protein storage vacuole.||[http://arabidopsis.org/servlets/TairObject?id=131459&type=locus TAIR]||Am, FPI||[http://www.uniprot.org/uniprot/Q84MI7 Grape] |
|- | |- | ||
| − | | TOC1||Pseudo response regulator involved in the generation of circadian rhythms. TOC1 appears to shorten the period of circumnutation speed. TOC1 contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. PRR3 may increase the stability of TOC1 by preventing interactions between TOC1 and the F-box protein ZTL. Expression of TOC1 is correlated with rhythmic changes in chromatin organization.||[http://arabidopsis.org/servlets/TairObject?id=133196&type=locus TAIR]||L | + | | TOC1||Pseudo response regulator involved in the generation of circadian rhythms. TOC1 appears to shorten the period of circumnutation speed. TOC1 contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. PRR3 may increase the stability of TOC1 by preventing interactions between TOC1 and the F-box protein ZTL. Expression of TOC1 is correlated with rhythmic changes in chromatin organization.||[http://arabidopsis.org/servlets/TairObject?id=133196&type=locus TAIR]||L|| |
|- | |- | ||
| − | | TOE1||Pathogenesis-related transcriptional factor/ERF, DNA-binding||[http://arabidopsis.org/servlets/TairObject?id=34025&type=locus TAIR]|| | + | | TOE1||Pathogenesis-related transcriptional factor/ERF, DNA-binding||[http://arabidopsis.org/servlets/TairObject?id=34025&type=locus TAIR]|||| |
|- | |- | ||
| − | | TOE2||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=133319&type=locus TAIR]|| | + | | TOE2||Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent||[http://arabidopsis.org/servlets/TairObject?id=133319&type=locus TAIR]|||| |
|- | |- | ||
| − | | TOE3||Pathogenesis-related transcriptional factor/ERF, DNA-binding||[http://arabidopsis.org/servlets/TairObject?id=132134&type=locus TAIR]|| | + | | TOE3||Pathogenesis-related transcriptional factor/ERF, DNA-binding||[http://arabidopsis.org/servlets/TairObject?id=132134&type=locus TAIR]|||| |
|- | |- | ||
| − | | TSF||Encodes a floral inducer that is a homolog of FT. Plants overexpressing this gene flower earlier than Col. Loss-of-function mutations flower later in short days. TSF and FT play overlapping roles in the promotion of flowering, with FT playing the dominant role.TSF sequences show extensive variation in different accessions and may contribute to quantitative variation in flowering time in these accessions. TSF has a complex pattern of spatial expression; it is expressed mainly in phloem and expression is regulated by daylength and vernalization.||[http://arabidopsis.org/servlets/TairObject?id=26553&type=locus TAIR]||Am, FPI | + | | TSF||Encodes a floral inducer that is a homolog of FT. Plants overexpressing this gene flower earlier than Col. Loss-of-function mutations flower later in short days. TSF and FT play overlapping roles in the promotion of flowering, with FT playing the dominant role.TSF sequences show extensive variation in different accessions and may contribute to quantitative variation in flowering time in these accessions. TSF has a complex pattern of spatial expression; it is expressed mainly in phloem and expression is regulated by daylength and vernalization.||[http://arabidopsis.org/servlets/TairObject?id=26553&type=locus TAIR]||Am, FPI|| |
|- | |- | ||
| − | | TT16||Encodes a MADS box protein. Regulates proanthocyanidin biosynthesis in the inner-most cell layer of the seed coat. Also controls cell shape of the inner-most cell layer of the seed coat. Also shown to be necessary for determining the identity of the endothelial layer within the ovule. Paralogous to GOA. Plays a maternal role in fertilization and seed development.||[http://arabidopsis.org/servlets/TairObject?id=133655&type=locus TAIR]|| | + | | TT16||Encodes a MADS box protein. Regulates proanthocyanidin biosynthesis in the inner-most cell layer of the seed coat. Also controls cell shape of the inner-most cell layer of the seed coat. Also shown to be necessary for determining the identity of the endothelial layer within the ovule. Paralogous to GOA. Plays a maternal role in fertilization and seed development.||[http://arabidopsis.org/servlets/TairObject?id=133655&type=locus TAIR]|||| |
|- | |- | ||
| − | | UFO||Required for the proper identity of the floral meristem. Involved in establishing the whorled pattern of floral organs, in the control of specification of the floral meristem, and in the activation of APETALA3 and PISTILLATA. UFO is found at the AP3 promoter in a LFY-dependent manner, suggesting that it works with LFY to regulate AP3 expression. UFO may also promote the ubiquitylation of LFY.||[http://arabidopsis.org/servlets/TairObject?id=28094&type=locus TAIR]|| | + | | UFO||Required for the proper identity of the floral meristem. Involved in establishing the whorled pattern of floral organs, in the control of specification of the floral meristem, and in the activation of APETALA3 and PISTILLATA. UFO is found at the AP3 promoter in a LFY-dependent manner, suggesting that it works with LFY to regulate AP3 expression. UFO may also promote the ubiquitylation of LFY.||[http://arabidopsis.org/servlets/TairObject?id=28094&type=locus TAIR]|||| |
|- | |- | ||
| − | | ULP1A*||Encodes a deSUMOylating enzyme. In vitro it has both peptidase activity and isopeptidase activity: it can cleave the C-terminal residues from SUMO to activate it for attachment to a target protein and it can also act on the isopeptide bond between SUMO and another protein. In vitro assays suggest that this enzyme is active against SUMO1 and SUMO2. It has weak activity with SUMO3 and cannot act on SUMO5. The N-terminal regulatory region of this protein is required for full activity.||[http://arabidopsis.org/servlets/TairObject?id=36190&type=locus TAIR]|| | + | | ULP1A*||Encodes a deSUMOylating enzyme. In vitro it has both peptidase activity and isopeptidase activity: it can cleave the C-terminal residues from SUMO to activate it for attachment to a target protein and it can also act on the isopeptide bond between SUMO and another protein. In vitro assays suggest that this enzyme is active against SUMO1 and SUMO2. It has weak activity with SUMO3 and cannot act on SUMO5. The N-terminal regulatory region of this protein is required for full activity.||[http://arabidopsis.org/servlets/TairObject?id=36190&type=locus TAIR]|||| |
|- | |- | ||
| − | | ULP1B*||Cysteine-type peptidase activity; proteolysis||[http://arabidopsis.org/servlets/TairObject?id=128314&type=locus TAIR]|| | + | | ULP1B*||Cysteine-type peptidase activity; proteolysis||[http://arabidopsis.org/servlets/TairObject?id=128314&type=locus TAIR]|||| |
|- | |- | ||
| − | | UVR3*||Required for photorepair of 6-4 photoproducts in Arabidopsis thaliana.||[http://arabidopsis.org/servlets/TairObject?id=38932&type=locus TAIR]||L | + | | UVR3*||Required for photorepair of 6-4 photoproducts in Arabidopsis thaliana.||[http://arabidopsis.org/servlets/TairObject?id=38932&type=locus TAIR]||L|| |
|- | |- | ||
| − | | VEL1||Encodes a protein with similarity to VRN5 and VIN3.Contains both a fibronectin III and PHD finger domain. VEL1 is a part of a polycomb repressive complex (PRC2) that is involved in epigenetic silencing of the FLC flowering locus.||[http://arabidopsis.org/servlets/TairObject?id=128580&type=locus TAIR]||Au, V | + | | VEL1||Encodes a protein with similarity to VRN5 and VIN3.Contains both a fibronectin III and PHD finger domain. VEL1 is a part of a polycomb repressive complex (PRC2) that is involved in epigenetic silencing of the FLC flowering locus.||[http://arabidopsis.org/servlets/TairObject?id=128580&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | VEL2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32230&type=locus TAIR]||Au | + | | VEL2||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32230&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | VEL3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32249&type=locus TAIR]||Au | + | | VEL3||Protein of unknown function||[http://arabidopsis.org/servlets/TairObject?id=32249&type=locus TAIR]||Au|| |
|- | |- | ||
| − | | VIN3||Encodes a plant homeodomain protein VERNALIZATION INSENSITIVE 3 (VIN3). In planta VIN3 and VRN2, VERNALIZATION 2, are part of a large protein complex that can include the polycomb group (PcG) proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF), and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization.||[http://arabidopsis.org/servlets/TairObject?id=134483&type=locus TAIR]||Au, V | + | | VIN3||Encodes a plant homeodomain protein VERNALIZATION INSENSITIVE 3 (VIN3). In planta VIN3 and VRN2, VERNALIZATION 2, are part of a large protein complex that can include the polycomb group (PcG) proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF), and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization.||[http://arabidopsis.org/servlets/TairObject?id=134483&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | VIP3||The protein is composed of repeats of WD motif which is involved in protein complex formation. The gene is involved in flower timing and flower development. This gene is predicted to encode a protein with a DWD motif. It can bind to DDB1a in Y2H assays, and DDB1b in co-IP assays, and may be involved in the formation of a CUL4-based E3 ubiquitin ligase. Loss of gene function leads to a redistribution of H3K4me3 and K3K36me2 modifications within genes but not a change in the overall abundance of these modifications within chromatin.||[http://arabidopsis.org/servlets/TairObject?id=127868&type=locus TAIR]|| | + | | VIP3||The protein is composed of repeats of WD motif which is involved in protein complex formation. The gene is involved in flower timing and flower development. This gene is predicted to encode a protein with a DWD motif. It can bind to DDB1a in Y2H assays, and DDB1b in co-IP assays, and may be involved in the formation of a CUL4-based E3 ubiquitin ligase. Loss of gene function leads to a redistribution of H3K4me3 and K3K36me2 modifications within genes but not a change in the overall abundance of these modifications within chromatin.||[http://arabidopsis.org/servlets/TairObject?id=127868&type=locus TAIR]|||| |
|- | |- | ||
| − | | VIP4||Encodes highly hydrophilic protein involved in positively regulating FLC expression. Mutants are early flowering and show a loss of FLC expression in the absence of cold.||[http://arabidopsis.org/servlets/TairObject?id=132654&type=locus TAIR]|| | + | | VIP4||Encodes highly hydrophilic protein involved in positively regulating FLC expression. Mutants are early flowering and show a loss of FLC expression in the absence of cold.||[http://arabidopsis.org/servlets/TairObject?id=132654&type=locus TAIR]|||| |
|- | |- | ||
| − | | VRN1||Required for vernalization. Essential for the complete repression of FLC in vernalized plants. Required for the methylation of histone H3||[http://arabidopsis.org/servlets/TairObject?id=37620&type=locus TAIR]||V | + | | VRN1||Required for vernalization. Essential for the complete repression of FLC in vernalized plants. Required for the methylation of histone H3||[http://arabidopsis.org/servlets/TairObject?id=37620&type=locus TAIR]||V|| |
|- | |- | ||
| − | | VRN2||The VERNALIZATION2 (VRN2) gene mediates vernalization and encodes a nuclear-localized zinc finger protein with similarity to Polycomb group (PcG) proteins of plants and animals. In wild-type Arabidopsis, vernalization results in the stable reduction of the levels of the floral repressor FLC. In vrn2 mutants, FLC expression is downregulated normally in response to vernalization, but instead of remaining low, FLC mRNA levels increase when plants are returned to normal temperatures. VRN2 maintains FLC repression after a cold treatment, serving as a mechanism for the cellular memory of vernalization. Required for complete repression of FLC. Required for the methylation of histone H3||[http://arabidopsis.org/servlets/TairObject?id=228106&type=locus TAIR]||Au, V | + | | VRN2||The VERNALIZATION2 (VRN2) gene mediates vernalization and encodes a nuclear-localized zinc finger protein with similarity to Polycomb group (PcG) proteins of plants and animals. In wild-type Arabidopsis, vernalization results in the stable reduction of the levels of the floral repressor FLC. In vrn2 mutants, FLC expression is downregulated normally in response to vernalization, but instead of remaining low, FLC mRNA levels increase when plants are returned to normal temperatures. VRN2 maintains FLC repression after a cold treatment, serving as a mechanism for the cellular memory of vernalization. Required for complete repression of FLC. Required for the methylation of histone H3||[http://arabidopsis.org/servlets/TairObject?id=228106&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | VRN5||Encodes Vernalization Insensitive 3-like 1 (VIL1). VIL1 is involved in the photoperiod and vernalization of Arabidopsis by regulating expression of the related floral repressors Flowering Locus C (FLC) and Flowering Locus M (FLM). VIL1, along with VIN3 (Vernalization Insensitive 3) is necessary for the chromatin modification to FLC and FLM.||[http://arabidopsis.org/servlets/TairObject?id=39196&type=locus TAIR]||Au, V | + | | VRN5||Encodes Vernalization Insensitive 3-like 1 (VIL1). VIL1 is involved in the photoperiod and vernalization of Arabidopsis by regulating expression of the related floral repressors Flowering Locus C (FLC) and Flowering Locus M (FLM). VIL1, along with VIN3 (Vernalization Insensitive 3) is necessary for the chromatin modification to FLC and FLM.||[http://arabidopsis.org/servlets/TairObject?id=39196&type=locus TAIR]||Au, V|| |
|- | |- | ||
| − | | WUS||Homeobox gene controlling the stem cell pool. Expressed in the stem cell organizing center of meristems. Required to keep the stem cells in an undifferentiated state. Regulation of WUS transcription is a central checkpoint in stem cell control. The size of the WUS expression domain controls the size of the stem cell population through WUS indirectly activating the expression of CLAVATA3 (CLV3) in the stem cells and CLV3 repressing WUS transcription through the CLV1 receptor kinase signaling pathway. Repression of WUS transcription through AGAMOUS (AG) activity controls stem cell activity in the determinate floral meristem. Binds to TAAT element core motif. WUS is also involved in cell differentiation during anther development.||[http://arabidopsis.org/servlets/TairObject?id=34708&type=locus TAIR]|| | + | | WUS||Homeobox gene controlling the stem cell pool. Expressed in the stem cell organizing center of meristems. Required to keep the stem cells in an undifferentiated state. Regulation of WUS transcription is a central checkpoint in stem cell control. The size of the WUS expression domain controls the size of the stem cell population through WUS indirectly activating the expression of CLAVATA3 (CLV3) in the stem cells and CLV3 repressing WUS transcription through the CLV1 receptor kinase signaling pathway. Repression of WUS transcription through AGAMOUS (AG) activity controls stem cell activity in the determinate floral meristem. Binds to TAAT element core motif. WUS is also involved in cell differentiation during anther development.||[http://arabidopsis.org/servlets/TairObject?id=34708&type=locus TAIR]||||[http://www.uniprot.org/uniprot/C3UZN8 Grape] |
|- | |- | ||
| − | | ZTL||Encodes clock-associated PAS protein ZTL; Also known as FKF1-like protein 2 or ADAGIO1(ADO1). A protein containing a PAS domain ZTL contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. ZTL is the F-box component of an SCF complex implicated in the degradation of TOC1.||[http://arabidopsis.org/servlets/TairObject?id=134481&type=locus TAIR]||L | + | | ZTL||Encodes clock-associated PAS protein ZTL; Also known as FKF1-like protein 2 or ADAGIO1(ADO1). A protein containing a PAS domain ZTL contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. ZTL is the F-box component of an SCF complex implicated in the degradation of TOC1.||[http://arabidopsis.org/servlets/TairObject?id=134481&type=locus TAIR]||L|| |
|} | |} | ||
Revision as of 18:57, 21 February 2013
Contents
To Do
Look up grape and strawberry orthologs of Arabidopsis genes using Mouhu et al., 2009, UniProt Grape, and UniProt Strawberry.
Prepare for presentation.
Aggregate all sequences into a single document in proper FASTA format.
Arabidopsis Flowering Pathway (2004-Present)
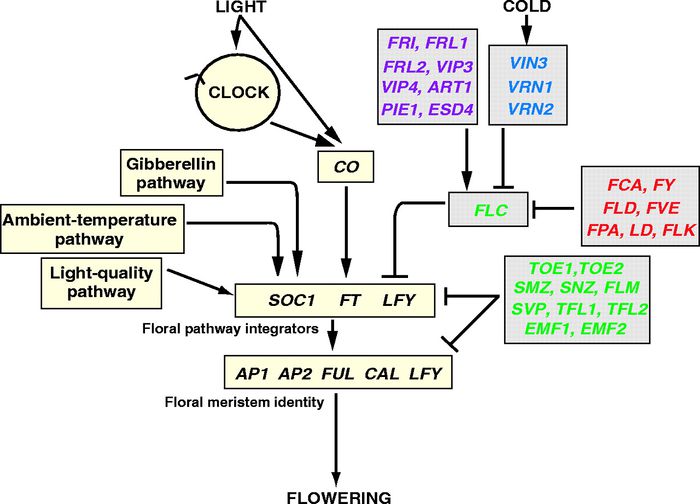
Overall pathway for the time of bloom from Henderson and Dean, 2004.
Overall pathway for the time of bloom from Schneitz, 2009.
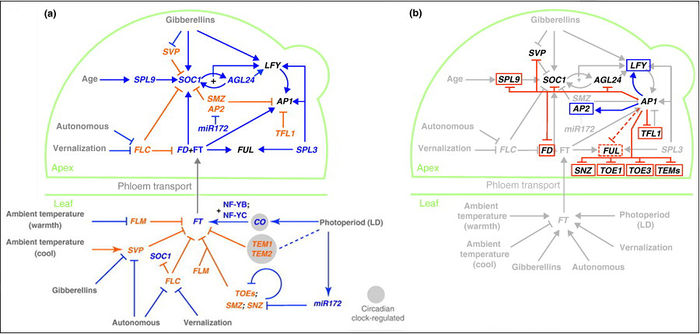
"The Arabidopsis floral transition network has been delineated as a result of extensive genetic analyses (depicted in a), but genome-wide studies have uncovered new molecular interactions and highlighted the role of AP1 as both a molecular switch and a network hub (shown in b)."
Overall pathway for the time of bloom from Wellmer and Riechmann, 2010 and Ferrier et al., 2011.
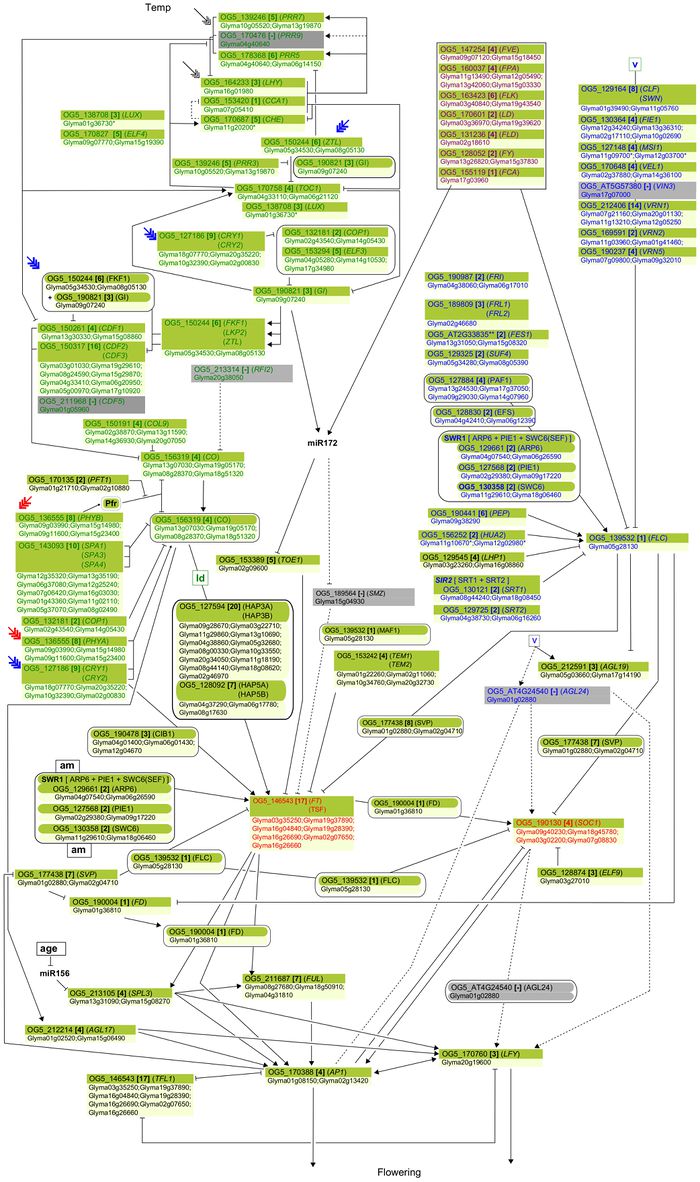
"Soybean genes marked with * are those that share the same clade with Arabidopsis genes but have not been investigated so far as flowering genes ... Arrows show promoting effects, T-bars show repressing effects. Environmental cues are shown as lower case letters in square boxes; ‘v’ is extended cold (vernalization); ‘ld’ is long days; ‘sd’ is short days; ‘am’ is ambient (non-vernalizing) temperature. Genes are shown in italics and proteins in non-italics in ovals. ‘Pfr’ indicates Pfr phytochrome signaling. Arabidopsis genes assigned to specific pathways are color-coded (photoperiod pathway in green, vernalization in blue and autonomous pathway in purple). Flowering pathway integrators are shown in red. Triple headed arrows indicate activation by red or blue light."
Overall pathway for the time of bloom from Jung et al., 2012.
All Arabidopsis Flowering Pathways
Henderson and Dean, 2004; Yamaguchi et al., 2005; Liu et al., 2009; Taiz and Zeiger, 2010; Boss et al., 2004; Zhang et al., 2011; Ballerini and Kramer, 2011; Posé et al., 2012; Kim and Sung, 2010; Wellmer and Riechmann, 2010; Farrona et al., 2008; He and Amasino, 2005; Max Planck Institute for Plant Breeding Research; Bäurle and Dean, 2006; Sung et al., 2003; Schneitz, 2009; Amasino, 2005; Ferrier et al., 2011; Izawa et al., 2003; Jung et al., 2012
Arabidopsis Flowering Genes
Note: All genes are compiled from Jung et al., 2012. Formatting is the same as Jung et al., 2012 where a * indicates 1 of "24 Arabidopsis genes, which had not been previously investigated for their roles in flowering, [that] belong to [ortholog groups] with known Arabidopsis flowering genes." Moreover, for pathways, "Am: Ambient temperature pathway, Au: Autonomous pathway, FPI: Flowering Pathway Intergrators, L: Light signaling pathway, MI: Meristem Identity, V: Vernalization pathway."
| Arabidopsis Gene | Function (All function information is directly copied from the respective AA Sequence page) | AA Sequence | Pathway | Potential Ortholog |
|---|---|---|---|---|
| ABF1 | Binds to the ABA-responsive element (ABRE). Could participate in abscisic acid-regulated gene expression. | UniProtKB | ||
| ABF2 | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | ||
| ABF3 | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. | UniProtKB | ||
| ABF4 | Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. | UniProtKB | ||
| ABI5 | Participates in ABA-regulated gene expression during seed development and subsequent vegetative stage by acting as the major mediator of ABA repression of growth. Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter and to the ABRE of the Em1 and Em6 genes promoters. Can also trans-activate its own promoter, suggesting that it is autoregulated. Plays a role in sugar-mediated senescence. | UniProtKB | ||
| AG | Probable transcription factor involved in the control of organ identity during the early development of flowers. Is required for normal development of stamens and carpels in the wild-type flower. Plays a role in maintaining the determinacy of the floral meristem. Acts as C class cadastral protein by repressing the A class floral homeotic genes like APETALA1. Forms a heterodimer via the K-box domain with either SEPALATTA1/AGL2, SEPALATTA2/AGL4, SEPALLATA3/AGL9 or AGL6 that could be involved in genes regulation during floral meristem development. | UniProtKB | Grape | |
| AGL14 | Probable transcription factor. | UniProtKB | V | |
| AGL15 | Transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Acts as both an activator and a repressor of transcription. Binds DNA in a sequence-specific manner in large CArG motif 5'-CC (A/T)8 GG-3'. Participates probably in the regulation of programs active during the early stages of embryo development. Prevents premature perianth senescence and abscission, fruits development and seed desiccation. Stimulates the expression of at least DTA4, LEC2, FUS3, ABI3, AT4G38680/CSP2 and GRP2B/CSP4. Can enhance somatic embryo development in vitro. | UniProtKB | ||
| AGL17 | Probable transcription factor. | UniProtKB | ||
| AGL18 | Probable transcription factor involved in the negative regulation of flowering, probably through the photoperiodic pathway. Prevents premature flowering. Downstream regulator of a subset of the MIKC* MADS-controlled genes required during pollen maturation. | UniProtKB | ||
| AGL19 | Probable transcription factor that promotes flowering, especially in response to vernalization by short periods of cold, in an FLC-inpedendent manner. | UniProtKB | V | |
| AGL21* | Probable transcription factor. | UniProtKB | ||
| AGL24 | Transcription activator that mediates floral transition in response to vernalization. Promotes inflorescence fate in apical meristems. Acts in a dosage-dependent manner. Probably involved in the transduction of RLK-mediated signaling (e.g. IMK3 pathway). Together with AP1 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. When associated with SOC1, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. Confers inflorescence characteristics to floral primordia and early flowering. | UniProtKB | V | |
| AGL31 | Probable transcription factor that prevents vernalization by short periods of cold. Acts as a floral repressor. | UniProtKB | Am, FPI, V | |
| AGL6 | Probable transcription factor. Forms a heterodimer via the K-box domain with AG, that could be involved in genes regulation during floral meristem development. | UniProtKB | ||
| AGL79 | - | UniProtKB | ||
| AP1 | Transcription factor that promotes early floral meristem identity in synergy with LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Is indispensable for normal development of sepals and petals in flowers. Regulates positively the B class homeotic proteins APETALA3 and PISTILLATA with the cooperation of LEAFY and UFO. Interacts with SEPALLATA3 or AP3/PI heterodimer to form complexes that could be involved in genes regulation during floral meristem development. Regulates positively AGAMOUS in cooperation with LEAFY. Displays a redundant function with CAULIFLOWER in the up-regulation of LEAFY. Together with AGL24 and SVP, controls the identity of the floral meristem and regulates expression of class B, C and E genes. Represses flowering time genes AGL24, SVP and SOC1 in emerging floral meristems. | UniProtKB | FPI, MI | Grape |
| AP2 | Probable transcriptional activator that promotes early floral meristem identity. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Plays a central role in the specification of floral identity, particularly for the normal development of sepals and petals in the wild-type flower. Acts as A class cadastral protein by repressing the C class floral homeotic gene AGAMOUS in association with other repressors like LEUNIG and SEUSS. It is also required during seed development. | UniProtKB | Grape | |
| AP3 | Probable transcription factor involved in the genetic control of flower development. Is required for normal development of petals and stamens in the wild-type flower. Forms a heterodimer with PISTILLATA that is required for autoregulation of both AP3 and PI genes. AP3/PI heterodimer interacts with APETALA1 or SEPALLATA3 to form a ternary complex that could be responsible for the regulation of the genes involved in the flower development. AP3/PI heterodimer activates the expression of NAP. | UniProtKB | Grape | |
| APRR3 | Controls photoperiodic flowering response. Component of the circadian clock. Controls the degradation of APRR1/TOC1 by the SCF(ZTL) complex. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L | |
| APRR5 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L | |
| APRR9 | Controls photoperiodic flowering response. Seems to be one of the component of the circadian clock. Expression of several members of the ARR-like family is controlled by circadian rhythm. The particular coordinated sequential expression of APRR9, APRR7, APRR5, APRR3 and APPR1 result to circadian waves that may be at the basis of the endogenous circadian clock. | UniProtKB | L | |
| AREB3 | Binds to the embryo specification element and the ABA-responsive element (ABRE) of the Dc3 gene promoter. Could participate in abscisic acid-regulated gene expression during seed development. | UniProtKB | ||
| AT1G26790 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. | UniProtKB | L | |
| AT1G29160 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. Acts as a negative regulator in the phytochrome-mediated light responses. Controls phyB-mediated end-of-day response and the phyA-mediated anthocyanin accumulation. | UniProtKB | L | |
| AT1G50680 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | ||
| AT1G51120 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | ||
| AT2G23070 | Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins. | UniProtKB | ||
| AT2G23080 | Casein kinases are operationally defined by their preferential utilization of acidic proteins such as caseins as substrates. The alpha chain contains the catalytic site. May act as an ectokinase that phosphorylates several extracellular proteins. | UniProtKB | ||
| AT2G25920 | Uncharacterized protein | UniProtKB | ||
| AT2G34140 | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. | UniProtKB | L | |
| AT2G46670* | Putative uncharacterized protein | UniProtKB | L | |
| AT3G21320 | Uncharacterized protein | UniProtKB | L | |
| AT3G25730 | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. | UniProtKB | L | |
| AT4G16810 | Putative uncharacterized protein | UniProtKB | ||
| AT5G08230* | Probable transcrition factor that seems to be involved in mRNA processing. | UniProtKB | V | |
| AT5G10625* | Modulates the competence to flowering of apical meristems. | UniProtKB | ||
| AT5G23280* | Transcription factor | UniProtKB | L | |
| AT5G27220 | Frigida-like protein | UniProtKB | ||
| AT5G42910 | Could participate in abscisic acid-regulated gene expression. | UniProtKB | ||
| AT5G44080 | Basic leucine zipper transcription factor | UniProtKB | ||
| AT5G48250* | Zinc finger protein Constans-like 10 | UniProtKB | L | |
| AT5G59570* | Putative transcription factor | UniProtKB | L | |
| AT5G62040 | May form complexes with phosphorylated ligands by interfering with kinases and their effectors. | UniProtKB | ||
| ATARP6 | Component of the SWR1 complex which mediates the ATP-dependent exchange of histone H2A for the H2A variant H2A.F/Z leading to transcriptional regulation of selected genes (e.g. FLC) by chromatin remodeling. Required for the activation of FLC and FLC/MAF genes expression to levels that inhibit flowering, through both histone H3 and H4 acetylation and methylation mechanisms. Involved in several developmental processes including organization of plant organs, leaves formation, flowering time repression, and fertility. Modulates photoperiod-dependent epidermal leaves cell development; promotes cell division in long days, and cell expansion/division in short days. May be involved in the regulation of pathogenesis-related proteins (PRs). | UniProtKB | Am, V | |
| ATBZIP27 | Transcription factor | GenScript | Am, MI | |
| ATC | May form complexes with phosphorylated ligands by interfering with kinases and their effectors. Can substitute for TERMINAL FLOWER 1 (in vitro). | UniProtKB | ||
| ATCOL4 | Sequence-specific DNA binding transcription factor activity, zinc ion binding. | TAIR | L | |
| ATCOL5 | Sequence-specific DNA binding transcription factor activity, zinc ion binding. | TAIR | L | |
| ATSWC6 | Encodes SERRATED LEAVES AND EARLY FLOWERING (SEF), an Arabidopsis homolog of the yeast SWC6 protein, a conserved subunit of the SWR1/SRCAP complex. SEF loss-of-function mutants have a pleiotropic phenotype characterized by serrated leaves, frequent absence of inflorescence internodes, bushy aspect, and flowers with altered number and size of organs. sef plants flower earlier than wild-type plants both under inductive and non-inductive photoperiods. SEF, ARP6 and PIE1 might form a molecular complex in Arabidopsis related to the SWR1/SRCAP complex identified in other eukaryotes. | TAIR | V | |
| CAL | Probable transcription factor that promotes early floral meristem identity in synergy with APETALA1, FRUITFULL and LEAFY. Is required subsequently for the transition of an inflorescence meristem into a floral meristem. Seems to be partially redundant to the function of APETALA1. | UniProtKB | FPI, MI | |
| CCA1 | Transcription factor involved in the circadian clock and in the phytochrome regulation. Binds to the promoter regions of APRR1/TOC1 and TCP21/CHE to repress their transcription. Binds to the promoter regions of CAB2A and CAB2B to promote their transcription. Represses both LHY and itself. | UniProtKB | L | |
| CDF1 | Cell growth defect factor | UniProtKB | L | |
| CDF2 | Cell growth defect factor | UniProtKB | L | |
| CDF3 | Plant non-specific lipid-transfer proteins transfer phospholipids as well as galactolipids across membranes. May play a role in wax or cutin deposition in the cell walls of expanding epidermal cells and certain secretory tissues. | UniProtKB | L | |
| CDF5 | Cell growth defect factor | - | L | |
| CHE | - | UniProtKB | L | |
| CIB1 | Encodes a transcription factor CIB1 (cryptochrome-interacting basic-helix-loop-helix). CIB1 interacts with CRY2 (cryptochrome 2) in a blue light-specific manner in yeast and Arabidopsis cells, and it acts together with additional CIB1-related proteins to promote CRY2-dependent floral initiation. CIB1 positively regulates FT expression. | TAIR | L | |
| CKA1 | Casein kinase II (CK2) catalytic subunit (alpha 1). One known substrate of CK2 is Phytochrome Interacting Factor 1 (PIF1). CK2-mediated phosphorylation enhances the light-induced degradation of PIF1 to promote photomorphogenesis. | TAIR | ||
| CKA2 | Encodes the casein kinase II (CK2) catalytic subunit (alpha). | TAIR | ||
| CLF | Similar to the product of the Polycomb-group gene Enhancer of zeste. Required for stable repression of AG and AP3. Putative role in cell fate determination. Involved in the control of leaf morphogenesis. mutants exhibit curled, involute leaves. AGAMOUS and APETALA3 are ectopically expressed in the mutant. | TAIR | Au, V | |
| CO | Encodes a protein showing similarities to zinc finger transcription factors, involved in regulation of flowering under long days. Acts upstream of FT and SOC1. | TAIR | FPI, L | |
| COL1 | Homologous to the flowering-time gene CONSTANS. | TAIR | FPI, L | |
| COL2 | Homologous to the flowering-time gene CONSTANS (CO) encoding zinc-finger proteins | TAIR | FPI, L | |
| COL3 | Positive regulator of photomorphogenesis that acts downstream of COP1 but can promote lateral root development independently of COP1 and also function as a daylength-sensitive regulator of shoot branching. | TAIR | ||
| COL9 | This gene belongs to the CO (CONSTANS) gene family. This gene family is divided in three subgroups: groups III, to which COL9 belongs, is characterised by one B-box (supposed to regulate protein-protein interactions) and a second diverged zinc finger. COL9 downregulates expression of CO (CONSTANS) as well as FT and SOC1 which are known regulatory targets of CO. | TAIR | L | |
| COP1 | Represses photomorphogenesis and induces skotomorphogenesis in the dark. Contains a ring finger zinc-binding motif, a coiled-coil domain, and several WD-40 repeats, similar to G-beta proteins. The C-terminus has homology to TAFII80, a subunit of the TFIID component of the RNA polymerase II of Drosophila. Nuclear localization in the dark and cytoplasmic in the light. | TAIR | L | |
| CRY1 | Encodes CRY1, a flavin-type blue-light photoreceptor with ATP binding and autophosphorylation activity. Functions in perception of blue / green ratio of light. The photoreceptor may be involved in electron transport. Mutant phenotype displays a blue light-dependent inhibition of hypocotyl elongation. Photoreceptor activity requires light-induced homodimerisation of the N-terminal CNT1 domains of CRY1. Involved in blue-light induced stomatal opening. The C-terminal domain of the protein undergoes a light dependent conformational change. Also involved in response to circadian rhythm. Mutants exhibit long hypocotyl under blue light and are out of phase in their response to circadian rhythm. CRY1 is present in the nucleus and cytoplasm. Different subcellular pools of CRY1 have different functions during photomorphogenesis of Arabidopsis seedlings. | TAIR | L | Grape |
| CRY2 | Blue light receptor mediating blue-light regulated cotyledon expansion and flowering time. Positive regulator of the flowering-time gene CONSTANS. This gene possesses a light-induced CNT2 N-terminal homodimerisation domain.Involved in blue-light induced stomatal opening. Involved in triggering chromatin decondensation. An 80-residue motif (NC80) is sufficient to confer CRY2's physiological function. It is proposed that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a "closed" conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an "open" conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses. Cry2 phosphorylation and degradation both occur in the nucleus. | TAIR | L | |
| DPBF2 | Basic leucine zipper transcription factor (BZIP67), identical to basic leucine zipper transcription factor GI:18656053 from (Arabidopsis thaliana); identical to cDNA basic leucine zipper transcription factor (atbzip67 gene) GI:18656052. Located in the nucleus and expressed during seed maturation in the cotyledons. | TAIR | ||
| E12A11 | Encodes a member of the FT and TFL1 family of phosphatidylethanolamine-binding proteins. It is expressed in seeds and up-regulated in response to ABA. Loss of function mutants show decreased rate of germination in the presence of ABA. ABA dependent regulation is mediated by both ABI3 and ABI5. ABI5 promotes MFT expression, primarily in the radicle-hypocotyl transition zone and ABI3 suppresses it in the seed. | TAIR | ||
| EEL | Transcription factor homologous to ABI5. Regulates AtEm1 expression by binding directly at the AtEm1 promoter. Located in the nucleus and expressed during seed maturation in the cotyledons and later in the whole embryo. | TAIR | ||
| EFS | Encodes a protein with histone lysine N-methyltransferase activity required specifically for the trimethylation of H3-K4 in FLC chromatin (and not in H3-K36 dimethylation). Acts as an inhibitor of flowering specifically involved in the autonomous promotion pathway. EFS also regulates the expression of genes involved in carotenoid biosynthesis.Modification of histone methylation at the CRTISO locus reduces transcript levels 90%. The increased shoot branching seen in some EFS mutants is likely due to the carotenoid biosynthesis defect having an effect on stringolactones.Required for ovule, embryo sac, anther and pollen development. | TAIR | V | |
| ELF3 | Encodes a nuclear protein that is expressed rhythmically and interacts with phytochrome B to control plant development and flowering through a signal transduction pathway. Required component of the core circadian clock regardless of light conditions. | TAIR | L | |
| ELF4 | Encodes a novel nuclear 111 amino-acid phytochrome-regulated component of a negative feedback loop involving the circadian clock central oscillator components CCA1 and LHY. ELF4 is necessary for light-induced expression of both CCA1 and LHY, and conversely, CCA1 and LHY act negatively on light-induced ELF4 expression. ELF4 promotes clock accuracy and is required for sustained rhythms in the absence of daily light/dark cycles. It is involved in the phyB-mediated constant red light induced seedling de-etiolation process and may function to coregulate the expression of a subset of phyB-regulated genes. | TAIR | L | |
| ELF4-L3 | Protein of unknown function | TAIR | ||
| ELF4-L2 | Protein of unknown function | TAIR | ||
| ELF4-L3 | Protein of unknown function | TAIR | L | |
| ELF4-L4 | Protein of unknown function | TAIR | ||
| ELF5 | Nuclear targeted protein involved in flowering time regulation that affects flowering time independent of FLC | TAIR | ||
| ELF6 | Early Flowering 6 (ELF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a repressor in the photoperiod pathway. ELF6 interacts with BES1 in a Y2H assay, in vitro, and in Arabidosis protoplasts (based on BiFC). ELF6 may play a role in brassinosteroid signaling by affecting histone methylation in the promoters of BR-responsive genes. | TAIR | ||
| ELF9 | Encodes a RNA binding protein ELF9 (EARLY FLOWERING9). Loss of ELF9 function in the Wassilewskija ecotype causes early flowering in short days. ELF9 reduces SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1) transcript levels, possibly via nonsense-mediated mRNA decay. | TAIR | FPI | |
| EMF1 | Involved in regulating reproductive development | TAIR | ||
| EMF2 | Polycomb group protein with zinc finger domain involved in negative regulation of reproductive development. Forms a complex with FIE, CLF, and MSI1. This complex modulates the expression of target genes including AG, PI and AP3. | TAIR | ||
| ESD4 | EARLY IN SHORT DAYS 4 Arabidopsis mutant shows extreme early flowering and alterations in shoot development. It encodes a SUMO protease, located predominantly at the periphery of the nucleus. Accelerates the transition from vegetative growth to flowering. Probably acts in the same pathway as NUA in affecting flowering time, vegetative and inflorescence development. | TAIR | ||
| FCA | Involved in the promotion of the transition of the vegetative meristem to reproductive development. Four forms of the protein (alpha, beta, delta and gamma) are produced by alternative splicing. Involved in RNA-mediated chromatin silencing. At one point it was believed to act as an abscisic acid receptor but the paper describing that function was retracted. | TAIR | Am, Au | Grape |
| FD | bZIP protein required for positive regulation of flowering. Mutants are late flowering. FD interacts with FT to promote flowering.Expressed in the shoot apex in floral anlagen, then declines in floral primordia. | TAIR | Am, MI | |
| FES1 | Encodes a zinc finger domain containing protein that is expressed in the shoot/root apex and vasculature, and acts with FRI to repress flowering.FES1 mutants in a Col(FRI+) background will flower early under inductive conditions. | TAIR | V | |
| FIE1 | Encodes a protein similar to the transcriptional regular of the animal Polycomb group and is involved in regulation of establishment of anterior-posterior polar axis in the endosperm and repression of flowering during vegetative phase. Mutation leads endosperm to develop in the absence of fertilization and flowers to form in seedlings and non-reproductive organs. Also exhibits maternal effect gametophytic lethal phenotype, which is suppressed by hypomethylation. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. In the ovule, the FIE transcript levels increase transiently just after fertilization. | TAIR | Au, V | |
| FIS2 | Encodes a negative regulator of seed development in the absence of pollination. In the ovule, the FIS2 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization. | TAIR | ||
| FKF1 | Encodes FKF1, a flavin-binding kelch repeat F box protein, is clock-controlled, regulates transition to flowering. Forms a complex with GI on the CO promoter to regulate CO expression. | TAIR | L | |
| FLC | MADS-box protein encoded by FLOWERING LOCUS C - transcription factor that functions as a repressor of floral transition and contributes to temperature compensation of the circadian clock. Expression is downregulated during cold treatment. Vernalization, FRI and the autonomous pathway all influence the state of FLC chromatin. Both maternal and paternal alleles are reset by vernalization, but their earliest activation differs in timing and location. Histone H3 trimethylation at lysine 4 and histone acetylation are associated with active FLC expression, whereas histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC repression. Expression is also repressed by two small RNAs (30- and 24-nt) complementary to the FLC sense strand 3’ to the polyA site. The small RNAs are most likely derived from an antisense transcript of FLC. Interacts with SOC1 and FT chromatin in vivo. Member of a protein complex. | TAIR | Am, FPI, V | Grape |
| FLD | Encodes a plant homolog of a SWIRM domain containing protein found in histone deacetylase complexes in mammals. Lesions in FLD result in hyperacetylation of histones in FLC chromatin, up-regulation of FLC expression and extremely delayed flowering. FLD plays a key role in regulating the reproductive competence of the shoot and results in different developmental phase transitions in Arabidopsis. | TAIR | Am, Au | |
| FLK | RNA binding, nucleic acid binding; positive regulation of flower development | TAIR | Au | |
| FLP1* | Encodes a small protein with unknown function and is similar to flower promoting factor 1. This gene is not expressed in apical meristem after floral induction but is expressed in roots, flowers, and in low abundance, leaves. | TAIR | ||
| FPA | FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. Double mutants with FCA have reduced fertility and single/double mutants have defects in siRNA mediated chromatin silencing. | TAIR | Au | |
| FPF1 | Encodes a small protein of 12.6 kDa that regulates flowering and is involved in gibberellin signalling pathway. It is expressed in apical meristems immediately after the photoperiodic induction of flowering. Genetic interactions with flowering time and floral organ identity genes suggest that this gene may be involved in modulating the competence to flower. There are two other genes similar to FPF1, FLP1 (At4g31380) and FLP2 (no locus name yet, on BAC F8F16 on chr 4). This is so far a plant-specific gene and is only found in long-day mustard, arabidopsis, and rice. | TAIR | ||
| FRI | Encodes a major determinant of natural variation in Arabidopsis flowering time. Dominant alleles of FRI confer a vernalization requirement causing plants to overwinter vegetatively. Many early flowering accessions carry loss-of-function fri alleles .Twenty distinct haplotypes that contain non-functional FRI alleles have been identified and the distribution analyzed in over 190 accessions. The common lab strains- Col and Ler each carry loss of function mutations in FRI. | TAIR | V | |
| FRL1 | Encodes a sterol-C24-methyltransferases involved in sterol biosynthesis. Mutants display altered sterol composition, serrated petals and sepals and altered cotyledon vascular patterning as well as ectopic endoreduplication. This suggests that suppression of endoreduplication is important for petal morphogenesis and that normal sterol composition is required for this suppression. | TAIR | V | |
| FRL2 | Family member of FRI-related genes that is required for the winter-annual habit. | TAIR | V | |
| FT | FT, together with LFY, promotes flowering and is antagonistic with its homologous gene, TERMINAL FLOWER1 (TFL1). FT is expressed in leaves and is induced by long day treatment. Either the FT mRNA or protein is translocated to the shoot apex where it induces its own expression. Recent data suggests that FT protein acts as a long-range signal. FT is a target of CO and acts upstream of SOC1. | TAIR | Am, FPI | Grape |
| FUL | MADS box gene negatively regulated by APETALA1 | TAIR | MI | Grape |
| FVE | Controls flowering. | TAIR | Am, Au | |
| FY | Encodes a protein with similarity to yeast Pfs2p, an mRNA processing factor. Involved in regulation of flowering time; affects FCA mRNA processing. Homozygous mutants are late flowering, null alleles are embryo lethal. | TAIR | Au | |
| GBF4 | Encodes a basic leucine zipper G-box binding factor that can bind to G-box motifs only as heterodimers with GBF2 or GBF3. A single amino acid change can confer G-box binding as homodimers. | TAIR | ||
| GI | Together with CONSTANTS (CO) and FLOWERING LOCUS T (FT), GIGANTEA promotes flowering under long days in a circadian clock-controlled flowering pathway. GI acts earlier than CO and FT in the pathway by increasing CO and FT mRNA abundance. Located in the nucleus. Regulates several developmental processes, including photoperiod-mediated flowering, phytochrome B signaling, circadian clock, carbohydrate metabolism, and cold stress response. The gene's transcription is controlled by the circadian clock and it is post-transcriptionally regulated by light and dark. Forms a complex with FKF1 on the CO promoter to regulate CO expression. | TAIR | L | |
| GRF1 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower | TAIR | FPI | |
| GRF10* | Encodes a 14-3-3 protein. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1 | TAIR | FPI | |
| GRF11* | Encodes a 14-3-3 protein. Binds H+-ATPase in response to blue light. | TAIR | FPI | |
| GRF12* | Encodes a 14-3-3 protein. | TAIR | FPI | |
| GRF2 | G-box binding factor GF14 omega encoding a 14-3-3 protein | TAIR | FPI | |
| GRF3 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Mutants result in smaller leaves indicating the role of the gene in leaf development. Expressed in root, shoot and flower. | TAIR | FPI | |
| GRF4 | GF14 protein phi chain member of 14-3-3 protein family. This protein is reported to interact with the BZR1 transcription factor involved in brassinosteroid signaling and may affect the nucleocytoplasmic shuttling of BZR1 | TAIR | FPI | |
| GRF5 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower. | TAIR | FPI | |
| GRF6 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in root, shoot and flower. | TAIR | FPI | |
| GRF7 | Encodes GF14 ν, a 14-3-3 protein isoform (14-3-3ν). | TAIR | FPI | |
| GRF8 | Growth regulating factor encoding transcription activator. One of the nine members of a GRF gene family, containing nuclear targeting domain. Involved in leaf development and expressed in shoot and flower. | TAIR | FPI | |
| HAP3A | Encodes a transcription factor from the nuclear factor Y (NF-Y) family, AtNF-YB1. Confers drought tolerance. | TAIR | FPI | |
| HAP3B | FPI | |||
| HAP5A | Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5A) and expressed in vegetative and reproductive tissues. | TAIR | FPI | |
| HAP5B | Encodes a protein with similarity to a subunit of the CCAAT promoter motif binding complex of yeast.One of two members of this class (HAP5B) and expressed in vegetative and reproductive tissues | TAIR | FPI | |
| HAP5C | Heme activated protein (HAP5c) | TAIR | FPI | |
| HUA2 | Putative transcription factor. Member of the floral homeotic AGAMOUS pathway.Mutations in HUA enhance the phenotype of mild ag-4 allele. Single hua mutants are early flowering and have reduced levels of FLC mRNA. Other MADS box flowering time genes such as FLM and MAF2 also appear to be regulated by HUA2. HUA2 normally activates FLC expression and enhances AG function. | TAIR | V | |
| LD | Encodes a nuclear localized protein with similarity to transcriptional regulators. Recessive mutants are late flowering. Expression of LFY is reduced in LD mutants. | TAIR | Au | |
| LDL1* | Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering time loci FLC and FWA. Located in nucleus. Negatively regulates root elongation. Involved in repression of LRP1 via histone deacetylation. | TAIR | Am, Au | |
| LDL2* | Encodes a homolog of human Lysine-Specific Demethylase1. Involved in H3K4 methylation of target genes including the flowering loci FLC and FWA. | TAIR | Am, Au | |
| LFY | Encodes transcriptional regulator that promotes the transition to flowering.Involved in floral meristem development. LFY is involved in the regulation of AP3 expression, and appears to bring the F-box protein UFO to the AP3 promoter. Amino acids 46-120 define a protein domain that mediates self-interaction. | TAIR | FPI, MI | Grape |
| LHP1 | Regulates the meristem response to light signals and the maintenance of inflorescence meristem identity. Influences developmental processes controlled by APETALA1. TFL2 silences specific genes within euchromatin but not genes positioned in heterochromatin. TFL2 protein localized preferentially to euchromatic regions and not to heterochromatic chromocenters. Involved in euchromatin organization. Required for epigenetic maintenance of the vernalized state. | TAIR | ||
| LHY | LHY encodes a myb-related putative transcription factor involved in circadian rhythm along with another myb transcription factor CCA1. | TAIR | L | |
| LKP2 | Encodes a member of F-box proteins that includes two other proteins in Arabidopsis (ZTL and FKF1). These proteins contain a unique structure containing a PAS domain at their N-terminus, an F-box motif, and 6 kelch repeats at their C-terminus. Overexpression results in arrhythmic phenotypes for a number of circadian clock outputs in both constant light and constant darkness, long hypocotyls under multiple fluences of both red and blue light, and a loss of photoperiodic control of flowering time. Although this the expression of this gene itself is not regulated by circadian clock, it physically interacts with Dof transcription factors that are transcriptionally regulated by circadian rhythm. LKP2 interacts with Di19, CO/COL family proteins. | TAIR | L | |
| LUX | Encodes a myb family transcription factor with a single Myb DNA-binding domain (type SHAQKYF) that is unique to plants and is essential for circadian rhythms, specifically for transcriptional regulation within the circadian clock. LUX is required for normal rhythmic expression of multiple clock outputs in both constant light and darkness. It is coregulated with TOC1 and seems to be repressed by CCA1 and LHY by direct binding of these proteins to the evening element in the LUX promoter. | TAIR | L | |
| MAF1 | MADS domain protein - flowering regulator that is closely related to FLC. Deletion of this locus in Nd ecotype is correlated with earlier flowering in short days suggesting function as a negative regulator of flowering. | TAIR | Am, FPI, V | |
| MAF3 | MADS domain protein - flowering regulator that is closely related to FLC | TAIR | Am, FPI, V | |
| MAF4 | Encodes MADS-box containing FLC paralog. Five splice variants have been identified but not characterized with respect to expression patterns and/or differing function. Overexpression of the gene in the Landsberg ecotype leads to a delay in flowering, transcript levels of MAF4 are reduced after a 6 week vernalization. | TAIR | Am, FPI, V | |
| MAF5 | Is upregulated during vernalization and regulates flowering time. Encodes MADS-domain protein. Two variants encoding proteins of 198 and 184 amino acids have been reported. | TAIR | Am, FPI, V | |
| MEA* | Encodes a putative transcription factor MEDEA (MEA) that negatively regulates seed development in the absence of fertilization. Mutations in this locus result in embryo lethality. MEA is a Polycomb group gene that is imprinted in the endosperm. The maternal allele is expressed and the paternal allele is silent. MEA is controlled by DEMETER (DME), a DNA glycosylase required to activate MEA expression, and METHYLTRANSFERASE I (MET1), which maintains CG methylation at the MEA locus. MEA is involved in the negative regulation of its own imprinted gene expression; the effect is not only allele-specific but also dynamically regulated during seed development. In the ovule, the MEA transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization | TAIR | Au, V | |
| MSI1 | Encodes a WD-40 repeat containing protein that functions in chromatin assembly as part of the CAF1 and FIE complex. Mutants exhibit parthenogenetic development that includes proliferation of unfertilized endosperm and embryos. In heterozygous plants 50% of embryos abort. Of the aborted embryos the early aborted class are homozygous and the later aborting lass are heterozygotes in which the defective allele is maternally transmitted. Other phenotypes include defects in ovule morphogenesis and organ initiation,as well as increased levels of heterochromatic DNA. MSI1 is needed for the transition to flowering. In Arabidopsis, the three CAF-1 subunits are encoded by FAS1, FAS2 and, most likely, MSI1, respectively. Mutations in FAS1 or FAS2 lead to increased frequency of homologous recombination and T-DNA integration in Arabidopsis. In the ovule, the MSI1 transcripts are accumulated at their highest level before fertilization and gradually decrease after fertilization. MSI is biallelically expressed, the paternall allele is expressed in the endosperm and embryo and is not imprinted. MSI1 forms a complex with RBR1 that is required for activation of the imprinted genes FIS2 and FWA. This activation is mediated by MSI1/RBR1 mediated repression of MET1. | TAIR | Au, V | |
| MSI2* | Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA | UniProtKB | Au, V | |
| MSI3* | Core histone-binding subunit that may target chromatin assembly factors, chromatin remodeling factors and histone deacetylases to their histone substrates in a manner that is regulated by nucleosomal DNA | UniProtKB | Au, V | |
| NF-YB10 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NF-YB3 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NF-YB7 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NF-YB8 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NF-YC3 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NF-YC4 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | FPI | |
| NFC5* | Cell cycle-related repressor genes encoding WD-repeat proteins. | TAIR | Am, Au | |
| PAF1 | Encodes a protein with extensive homology to the largest subunit of the multicatalytic proteinase complex (proteasome). Negatively regulates thiol biosynthesis and arsenic tolerance. | TAIR | V | |
| PAF2 | Encodes 20S proteasome subunit PAF2 (PAF2). | TAIR | V | |
| PEP | Encodes a novel Arabidopsis gene encoding a polypeptide with K-homology (KH) RNA-binding modules, which acts on vegetative growth and pistil development. Genetic studies suggest that PEP interacts with element(s) of the CLAVATA signaling pathway. | TAIR | V | |
| PFT1 | Encodes a nuclear protein that acts in a phyB pathway (but downstream of phyB) and induces flowering in response to suboptimal light conditions. PFT1 promotes flowering in CO dependent and independent pathways and integrates several environmental stimuli, such as light quality and JA-dependent defenses. Mutants are hypo-responsive to far-red and hyper-responsive to red light and flower late under long day conditions. Also shown to be a Mediator subunit regulating jasmonate-dependent defense. | TAIR | ||
| PHYA | Light-labile cytoplasmic red/far-red light photoreceptor involved in the regulation of photomorphogenesis. It exists in two inter-convertible forms: Pr and Pfr (active) and functions as a dimer.The N terminus carries a single tetrapyrrole chromophore, and the C terminus is involved in dimerization. It is the sole photoreceptor mediating the FR high irradiance response (HIR). Major regulator in red-light induction of phototropic enhancement. Involved in the regulation of de-etiolation. Involved in gravitropism and phototropism. Requires FHY1 for nuclear accumulation. | TAIR | L | Grape |
| PHYB | Red/far-red photoreceptor involved in the regulation of de-etiolation. Exists in two inter-convertible forms: Pr and Pfr (active). Involved in the light-promotion of seed germination and in the shade avoidance response. Promotes seedling etiolation in both the presence and absence of phytochrome A. Overexpression results in etiolation under far-red light. Accumulates in the nucleus after exposure to far red light. | TAIR | L | Grape |
| PHYC | Encodes the apoprotein of phytochrome;one of a family of photoreceptors that modulate plant growth and development. | TAIR | L | Grape |
| PHYD | Encodes a phytochrome photoreceptor with a function similar to that of phyB that absorbs the red/far-red part of the light spectrum and is involved in light responses. It cannot compensate for phyB loss in Arabidopsis but can substitute for tobacco phyB in vivo. | TAIR | L | |
| PHYE | Histidine kinase | TAIR | L | Grape |
| PI | Floral homeotic gene encoding a MADS domain transcription factor. Required for the specification of petal and stamen identities. | TAIR | Grape | |
| PIE1 | Encodes a protein similar to ATP-dependent, chromatin-remodeling proteins of the ISWI and SWI2/SNF2 family. Genetic analyses suggest that this gene is involved in multiple flowering pathways. Mutations in PIE1 results in suppression of FLC-mediated delay of flowering and causes early flowering in noninductive photoperiods independently of FLC. PIE1 is required for expression of FLC in the shoot apex but not in the root.Along with ARP6 forms a complex to deposit modified histone H2A.Z at several loci within the genome. This modification alters the expression of the target genes (i.e. FLC, MAF4, MAF6). | TAIR | V | |
| PIF3 | Transcription factor interacting with photoreceptors phyA and phyB. Forms a ternary complex in vitro with G-box element of the promoters of LHY, CCA1. Acts as a negative regulator of phyB signalling. It degrades rapidly after irradiation of dark grown seedlings in a process controlled by phytochromes. Does not play a significant role in controlling light input and function of the circadian clockwork. Binds to G- and E-boxes, but not to other ACEs. Binds to anthocyanin biosynthetic genes in a light- and HY5-independent fashion. PIF3 function as a transcriptional activator can be functionally and mechanistically separated from its role in repression of PhyB mediated processes. | TAIR | ||
| PRR7 | PRR7 and PRR9 are partially redundant essential components of a temperature-sensitive circadian system. CCA1 and LHY had a positive effect on PRR7 expression levels. Acts as transcriptional repressor of CCA1 and LHY. | TAIR | L | |
| RAV1 | Encodes an AP2/B3 domain transcription factor which is upregulated in response to low temperature. It contains a B3 DNA binding domain. It has circadian regulation and may function as a negative growth regulator. | TAIR | L | |
| REF6 | Relative of Early Flowering 6 (REF6) encodes a Jumonji N/C and zinc finger domain-containing protein that acts as a positive regulator of flowering in an FLC-dependent pathway. REF6 mutants have hyperacetylation of histone H4 at the FLC locus. REF6 interacts with BES1 in a Y2H assay and in vitro. REF6 may play a role in brassinoteroid signaling by affecting histone methylation in the promoters of BR-responsive genes. It is most closely related to the JHDM3 subfamily of JmjN/C proteins. | TAIR | ||
| RFI2 | Zinc ion binding | TAIR | L | |
| SEC* | Has O-linked N-acetyl glucosamine transferase activity. Similar to Arabidopsis SPY gene. | TAIR | ||
| SEP1 | Encodes a stress enhanced protein that localizes to the thylakoid membrane and whose mRNA is upregulated in response to high light intensity. It may be involved in chlorophyll binding. | TAIR | ||
| SEP2 | Stress enhanced protein 2 (SEP2) chlorophyll a/b-binding protein | TAIR | ||
| SEP3 | Member of the MADs box transcription factor family. SEP3 is redundant with SEP1 and 2. Flowers of SEP1/2/3 triple mutants show a conversion of petals and stamens to sepals.SEP3 forms heterotetrameric complexes with other MADS box family members and binds to the CArG box motif. | TAIR | ||
| SEP4 | This gene belongs to the family of SEP genes. It is involved in the development of sepals, petals, stamens and carpels. Additionally, it plays a central role in the determination of flower meristem and organ identity. | TAIR | ||
| SHP1* | One of two genes (SHP1 and SHP2) that are required for fruit dehiscence. The two genes control dehiscence zone differentiation and promote the lignification of adjacent cells. | TAIR | ||
| SHP2* | AGAMOUS [AG]-like MADS box protein (AGL5) involved in fruit development (valve margin and dehiscence zone differentiation). A putative direct target of AG. SHP2 has been shown to be a downstream gene of the complex formed by AG and SEP proteins (SEP4 alone does not form a functional complex with AG). | TAIR | ||
| SMZ | Encodes a AP2 domain transcription factor that can repress flowering. SMZ and its paralogous gene, SNARCHZAPFEN (SNZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering. | TAIR | ||
| SNZ | Encodes a AP2 domain transcription factor that can repress flowering. SNZ and its paralogous gene, SCHLAFMUTZE (SMZ), share a signature with partial complementarity to the miR172 microRNA, whose precursor is induced upon flowering. | TAIR | ||
| SOC1 | Transcription activator active in flowering time control. May integrate signals from the photoperiod, vernalization and autonomous floral induction pathways. Can modulate class B and C homeotic genes expression. When associated with AGL24, mediates effect of gibberellins on flowering under short-day conditions, and regulates the expression of LEAFY (LFY), which links floral induction and floral development. | UniProtKB | FPI | Grape |
| SPA1 | Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA1 is a PHYA signaling intermediate, putative regulator of PHYA signaling pathway. Light responsive repressor of photomorphogenesis. Involved in regulating circadian rhythms and flowering time in plants. Under constant light, the abundance of SPA1 protein exhibited circadian regulation, whereas under constant darkness, SPA1 protein levels remained unchanged. In addition, the spa1-3 mutation slightly shortened circadian period of CCA1, TOC1/PRR1 and SPA1 transcript accumulation under constant light. | TAIR | L | |
| SPA2 | Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA2 primarily regulates seedling development in darkness and has little function in light-grown seedlings or adult plants. | TAIR | L | |
| SPA3 | Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA3 (and SPA4) predominantly regulates elongation growth in adult plants. | TAIR | L | |
| SPA4 | Encodes a member of the SPA (suppressor of phyA-105) protein family (SPA1-SPA4). SPA proteins contain an N-terminal serine/threonine kinase-like motif followed by a coiled-coil structure and a C-terminal WD-repeat domain. SPA proteins function redundantly in suppressing photomorphogenesis in dark- and light-grown seedlings. SPA4 (and SPA3) predominantly regulates elongation growth in adult plants. | TAIR | L | |
| SPL3 | Encodes a member of the SPL (squamosa-promoter binding protein-like)gene family, a novel gene family encoding DNA binding proteins and putative transcription factors. Contains the SBP-box, which encodes the SBP-domain, required and sufficient for interaction with DNA. It binds DNA, may directly regulate AP1, and is involved in regulation of flowering and vegetative phase change. Its temporal expression is regulated by the microRNA miR156. The target site for the microRNA is in the 3'UTR. | TAIR | MI | |
| SPY | Encodes a N-acetyl glucosamine transferase that may glycosylate other molecules involved in GA signaling. Contains a tetratricopeptide repeat region, and a novel carboxy-terminal region. SPY acts as both a repressor of GA responses and as a positive regulation of cytokinin signalling. SPY may be involved in reducing ROS accumulation in response to stress. | TAIR | ||
| STK* | Encodes a MADS box transcription factor expressed in the carpel and ovules. Plays a maternal role in fertilization and seed development. | TAIR | ||
| STM | Class I knotted-like homeodomain protein that is required for shoot apical meristem (SAM) formation during embryogenesis and for SAM function throughout the lifetime of the plant. Functions by preventing incorporation of cells in the meristem center into differentiating organ primordia. | TAIR | ||
| SUF4 | Encodes SUF4 (SUPPRESSOR of FRI 4), a putative zinc-finger-containing transcription factor that is required for delayed flowering in winter-annual Arabidopsis. suf4 mutations strongly suppress the late-flowering phenotype of FRI (FRIGIDA) mutants. suf4 mutants also show reduced H3K4 trimethylation at FLC (FLOWERING LOCUS C), a floral inhibitor. SUF4 may act to specifically recruit a putative histone H3 methyltransferase EFS (EARLY FLOWERING IN SHORT DAYS) and the PAF1-like complex to the FLC locus. | TAIR | V | |
| SVP | Encodes a nuclear protein that acts as a floral repressor and that functions within the thermosensory pathway. SVP represses FT expression via direct binding to the vCArG III motif in the FT promoter. | TAIR | Am, V | Grape |
| SWN | Encodes a polycomb group protein. Forms part of a large protein complex that can include VRN2 (VERNALIZATION 2), VIN3 (VERNALIZATION INSENSITIVE 3) and polycomb group proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and CURLY LEAF (CLF). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. Performs a partially redundant role to MEA in controlling seed initiation by helping to suppress central cell nucleus endosperm proliferation within the FG. | TAIR | Au, V | |
| TEM1 | Encodes a member of the RAV transcription factor family that contains AP2 and B3 binding domains. Involved in the regulation of flowering under long days. Loss of function results in early flowering. Overexpression causes late flowering and repression of expression of FT. Novel transcriptional regulator involved in ethylene signaling. Promoter bound by EIN3. EDF1 in turn, binds to promoter elements in ethylene responsive genes. | TAIR | L | |
| TEM2 | Rav2 is part of a complex that has been named `regulator of the (H+)-ATPase of the vacuolar and endosomal membranes' (RAVE) | TAIR | L | |
| TFL1 | Controls inflorescence meristem identity. Involved in the floral initiation process. Ortholog of the Antirrhinum gene CENTRORADIALIS (CEN). Involved in protein trafficking to the protein storage vacuole. | TAIR | Am, FPI | Grape |
| TOC1 | Pseudo response regulator involved in the generation of circadian rhythms. TOC1 appears to shorten the period of circumnutation speed. TOC1 contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. PRR3 may increase the stability of TOC1 by preventing interactions between TOC1 and the F-box protein ZTL. Expression of TOC1 is correlated with rhythmic changes in chromatin organization. | TAIR | L | |
| TOE1 | Pathogenesis-related transcriptional factor/ERF, DNA-binding | TAIR | ||
| TOE2 | Sequence-specific DNA binding transcription factor activity; regulation of transcription, DNA-dependent | TAIR | ||
| TOE3 | Pathogenesis-related transcriptional factor/ERF, DNA-binding | TAIR | ||
| TSF | Encodes a floral inducer that is a homolog of FT. Plants overexpressing this gene flower earlier than Col. Loss-of-function mutations flower later in short days. TSF and FT play overlapping roles in the promotion of flowering, with FT playing the dominant role.TSF sequences show extensive variation in different accessions and may contribute to quantitative variation in flowering time in these accessions. TSF has a complex pattern of spatial expression; it is expressed mainly in phloem and expression is regulated by daylength and vernalization. | TAIR | Am, FPI | |
| TT16 | Encodes a MADS box protein. Regulates proanthocyanidin biosynthesis in the inner-most cell layer of the seed coat. Also controls cell shape of the inner-most cell layer of the seed coat. Also shown to be necessary for determining the identity of the endothelial layer within the ovule. Paralogous to GOA. Plays a maternal role in fertilization and seed development. | TAIR | ||
| UFO | Required for the proper identity of the floral meristem. Involved in establishing the whorled pattern of floral organs, in the control of specification of the floral meristem, and in the activation of APETALA3 and PISTILLATA. UFO is found at the AP3 promoter in a LFY-dependent manner, suggesting that it works with LFY to regulate AP3 expression. UFO may also promote the ubiquitylation of LFY. | TAIR | ||
| ULP1A* | Encodes a deSUMOylating enzyme. In vitro it has both peptidase activity and isopeptidase activity: it can cleave the C-terminal residues from SUMO to activate it for attachment to a target protein and it can also act on the isopeptide bond between SUMO and another protein. In vitro assays suggest that this enzyme is active against SUMO1 and SUMO2. It has weak activity with SUMO3 and cannot act on SUMO5. The N-terminal regulatory region of this protein is required for full activity. | TAIR | ||
| ULP1B* | Cysteine-type peptidase activity; proteolysis | TAIR | ||
| UVR3* | Required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. | TAIR | L | |
| VEL1 | Encodes a protein with similarity to VRN5 and VIN3.Contains both a fibronectin III and PHD finger domain. VEL1 is a part of a polycomb repressive complex (PRC2) that is involved in epigenetic silencing of the FLC flowering locus. | TAIR | Au, V | |
| VEL2 | Protein of unknown function | TAIR | Au | |
| VEL3 | Protein of unknown function | TAIR | Au | |
| VIN3 | Encodes a plant homeodomain protein VERNALIZATION INSENSITIVE 3 (VIN3). In planta VIN3 and VRN2, VERNALIZATION 2, are part of a large protein complex that can include the polycomb group (PcG) proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF), and SWINGER (SWN or EZA1). The complex has a role in establishing FLC (FLOWERING LOCUS C) repression during vernalization. | TAIR | Au, V | |
| VIP3 | The protein is composed of repeats of WD motif which is involved in protein complex formation. The gene is involved in flower timing and flower development. This gene is predicted to encode a protein with a DWD motif. It can bind to DDB1a in Y2H assays, and DDB1b in co-IP assays, and may be involved in the formation of a CUL4-based E3 ubiquitin ligase. Loss of gene function leads to a redistribution of H3K4me3 and K3K36me2 modifications within genes but not a change in the overall abundance of these modifications within chromatin. | TAIR | ||
| VIP4 | Encodes highly hydrophilic protein involved in positively regulating FLC expression. Mutants are early flowering and show a loss of FLC expression in the absence of cold. | TAIR | ||
| VRN1 | Required for vernalization. Essential for the complete repression of FLC in vernalized plants. Required for the methylation of histone H3 | TAIR | V | |
| VRN2 | The VERNALIZATION2 (VRN2) gene mediates vernalization and encodes a nuclear-localized zinc finger protein with similarity to Polycomb group (PcG) proteins of plants and animals. In wild-type Arabidopsis, vernalization results in the stable reduction of the levels of the floral repressor FLC. In vrn2 mutants, FLC expression is downregulated normally in response to vernalization, but instead of remaining low, FLC mRNA levels increase when plants are returned to normal temperatures. VRN2 maintains FLC repression after a cold treatment, serving as a mechanism for the cellular memory of vernalization. Required for complete repression of FLC. Required for the methylation of histone H3 | TAIR | Au, V | |
| VRN5 | Encodes Vernalization Insensitive 3-like 1 (VIL1). VIL1 is involved in the photoperiod and vernalization of Arabidopsis by regulating expression of the related floral repressors Flowering Locus C (FLC) and Flowering Locus M (FLM). VIL1, along with VIN3 (Vernalization Insensitive 3) is necessary for the chromatin modification to FLC and FLM. | TAIR | Au, V | |
| WUS | Homeobox gene controlling the stem cell pool. Expressed in the stem cell organizing center of meristems. Required to keep the stem cells in an undifferentiated state. Regulation of WUS transcription is a central checkpoint in stem cell control. The size of the WUS expression domain controls the size of the stem cell population through WUS indirectly activating the expression of CLAVATA3 (CLV3) in the stem cells and CLV3 repressing WUS transcription through the CLV1 receptor kinase signaling pathway. Repression of WUS transcription through AGAMOUS (AG) activity controls stem cell activity in the determinate floral meristem. Binds to TAAT element core motif. WUS is also involved in cell differentiation during anther development. | TAIR | Grape | |
| ZTL | Encodes clock-associated PAS protein ZTL; Also known as FKF1-like protein 2 or ADAGIO1(ADO1). A protein containing a PAS domain ZTL contributes to the plant fitness (carbon fixation, biomass) by influencing the circadian clock period. ZTL is the F-box component of an SCF complex implicated in the degradation of TOC1. | TAIR | L |