Difference between revisions of "Present Hybrid Promoter Designs cartoon fashion (3 major different types)"
| Line 11: | Line 11: | ||
[[Oligos_to_Build]]: Sequences we will need to make this XOR gate. | [[Oligos_to_Build]]: Sequences we will need to make this XOR gate. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== Missouri Western XOR Biological Design 1== | == Missouri Western XOR Biological Design 1== | ||
| Line 31: | Line 16: | ||
These two XOR circuits are designed to complement each other. Each recieves a cell-to-cell signal (AI-1 or AI-2) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce AI-2. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate. | These two XOR circuits are designed to complement each other. Each recieves a cell-to-cell signal (AI-1 or AI-2) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce AI-2. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
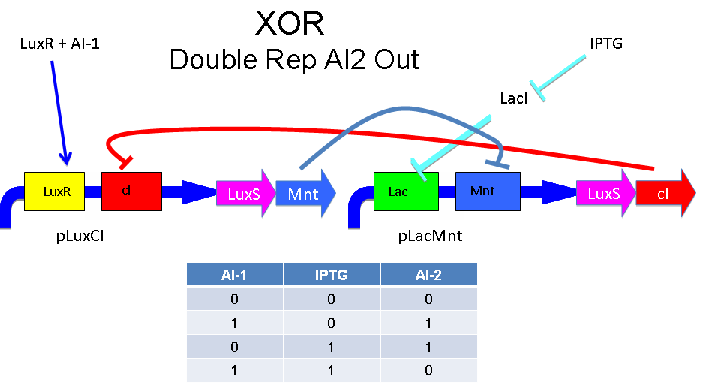
[[Image:XOR DR AI2.PNG]] | [[Image:XOR DR AI2.PNG]] | ||
Latest revision as of 14:58, 28 October 2008
Davidson XOR Biological Design
List of auto-inducers and their catalog numbers.
Here is an idea Malcolm and Laurie developed.
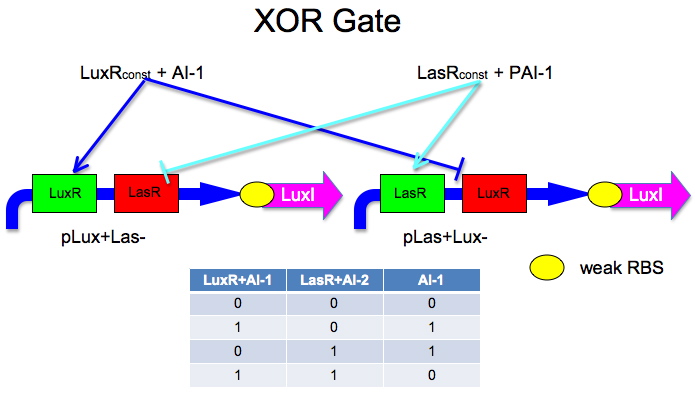
The idea is to have two mirrored halves of the system. LasR is regulated by PAI-1 {3-oxododecanoyl-HSL (3OC12HSL)} and LuxR is activated by AI-1 {3-oxohexanoyl-homoserine lactone (3OC6HSL)}. There is a potential problem in that the Lux half is more likely to get positive feedback than the Las half. This MAY not be a problem because 0/0 is leaky so we put a weak RBS to minimize leaky protein production. Also, if we add AI-2 and AI-1 is produced by leak, then the entire system shuts down. The repressor site is located between -35 and -10 of the promoter. The activator binding site is upstream of -35. This has been documented by Egland and Greenberg
Oligos_to_Build: Sequences we will need to make this XOR gate.
Missouri Western XOR Biological Design 1
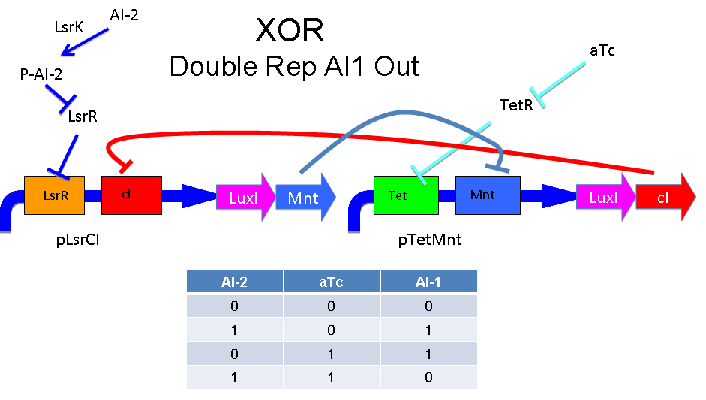
These two XOR circuits are designed to complement each other. Each recieves a cell-to-cell signal (AI-1 or AI-2) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce AI-2. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate.
Above - Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed.
Above - Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.