Difference between revisions of "Aaron D"
m |
|||
| (15 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
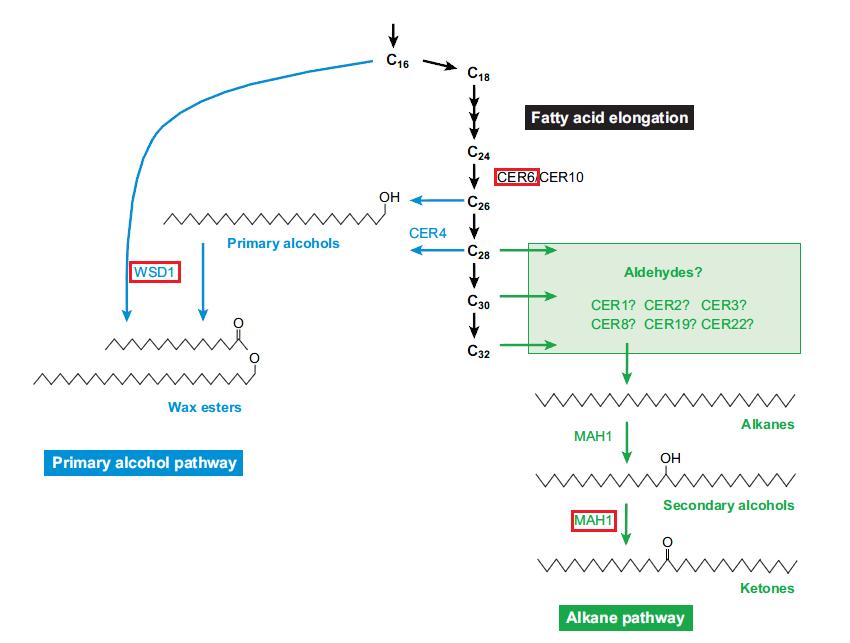
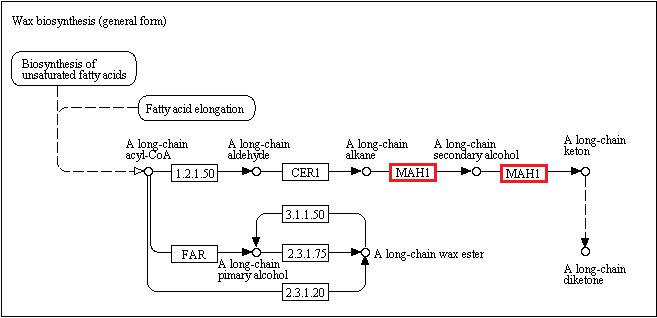
| − | Samuels et al. ([[Media:Samuels_color.pdf]]) provided a thorough analysis of wax production in ''Arabidopsis''. Additionally, pathways presented within the paper indicated genes involved specifically in ketone production. Based on the relatedness of two genera, I searched for orthologs of ''Arabidopsis'' within the ''Vaccinium'' scaffold collection. Genes were chosen to represent a variety of roles within wax synthesis and secretion. | + | Samuels et al. ([[Media:Samuels_color.pdf]]) provided a thorough analysis of wax production in ''Arabidopsis''. Additionally, pathways presented within the paper and the [http://www.genome.jp/kegg-bin/show_pathway?map00073 KEGG website] indicated genes involved specifically in ketone production. Based on the relatedness of two genera, I searched for orthologs of ''Arabidopsis'' within the ''Vaccinium'' scaffold collection. Genes were chosen to represent a variety of roles within wax synthesis and secretion. |
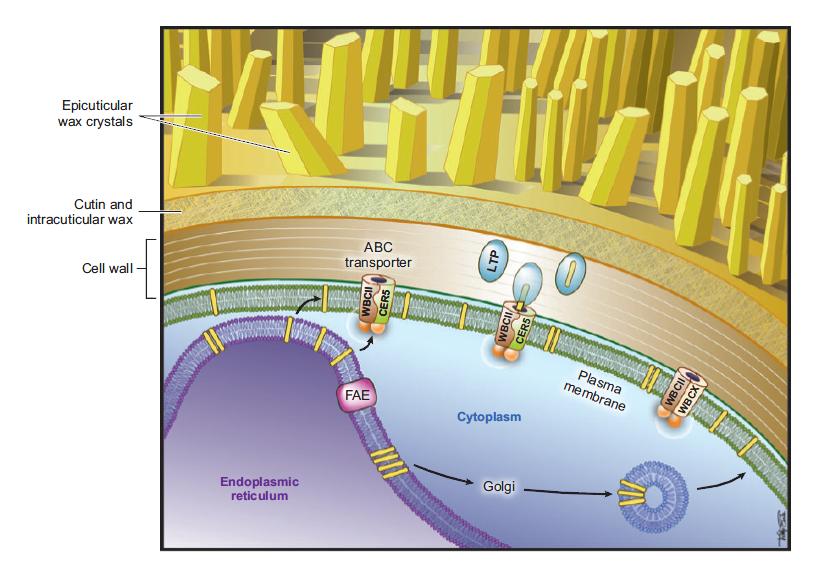
| − | Target genes: '' | + | Target genes: ''WBCII'' (transporter: wax secretion); ''CER6'' (fatty acid elongation); ''WSD'' (wax production); ''FATB'' (wax production); ''MAH1'' (ketone production) |
| Line 28: | Line 28: | ||
| − | ''' | + | '''Primers''' |
| − | '' | + | ''WBCII'' |
| Line 109: | Line 109: | ||
2. Forward Primer CTCTTCTCCCCCTTTCATACCT & Reverse Primer CTTCCTGGTGAAGAGAAATTGG (ta) x5 PCR product = 225 (230813 - 230822 bp) | 2. Forward Primer CTCTTCTCCCCCTTTCATACCT & Reverse Primer CTTCCTGGTGAAGAGAAATTGG (ta) x5 PCR product = 225 (230813 - 230822 bp) | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | '''ESTs''' | ||
| + | |||
| + | |||
| + | Expressed sequence tags (ESTs) were found using tBLASTn against the EST database of the [http://dev.vaccinium.org/tools/blast ''Vaccinium''] website. Queries were generated by copying the database sequence from a section of amino acids aligning with the target gene. Confirmation of any EST hits with a sufficient E-score was made using the [http://dev.vaccinium.org/unigene_search ''Unigene search''] feature of the Vaccinium website. | ||
| + | |||
| + | |||
| + | No ESTs were found for WSD or FATB. | ||
| + | |||
| + | |||
| + | '''''WBCII''''' | ||
| + | |||
| + | |||
| + | Scaffold 01191: DR067855 | ||
| + | |||
| + | [[File:DR067855.jpg]] | ||
| + | |||
| + | |||
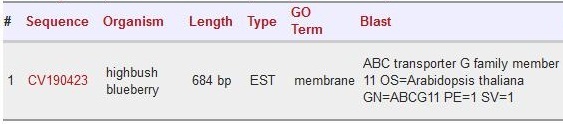
| + | Scaffold 01191: CV190423 | ||
| + | |||
| + | [[File:CV190423.jpg]] | ||
| + | |||
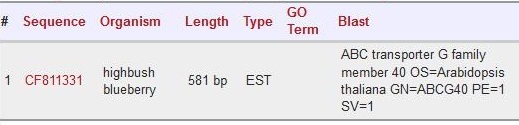
| + | |||
| + | Scaffold 01191: CF811331 | ||
| + | |||
| + | [[File:CF811331.jpg]] | ||
| + | |||
| + | |||
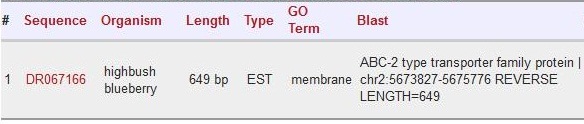
| + | Scaffold 00913: DR067166 | ||
| + | |||
| + | [[File:DR067166.jpg]] | ||
| + | |||
| + | |||
| + | '''''CER6''''' | ||
| + | |||
| + | |||
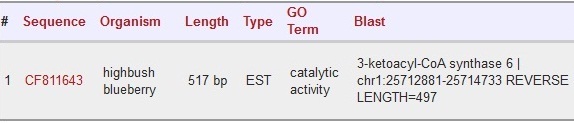
| + | Scaffold 00137 and 00268: CF811643 | ||
| + | |||
| + | [[File:CF811643.jpg]] | ||
| + | |||
| + | |||
| + | '''''MAH1''''' | ||
| + | |||
| + | (Cytochrome P450 listed as alternate name for ''MAH1'' on [http://www.arabidopsis.org The Arabidopsis Information Resource]) | ||
| + | |||
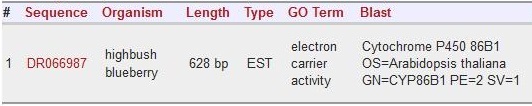
| + | Scaffold 01091: DR066987 | ||
| + | |||
| + | [[File:DR066987.jpg]] | ||
| + | |||
| + | |||
| + | Scaffold 00236: CV090972 | ||
| + | |||
| + | [[File:CV090972.jpg]] | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
| + | '''Scaffolds found using ESTs''' | ||
| + | |||
| + | |||
| + | The following represent scaffolds that may indicate paralogs of each gene within the blueberry genome. They were found by submitting the FASTA of the EST into a tBLASTx against the ''Vaccinium'' scaffold database. | ||
| + | |||
| + | |||
| + | '''''WBCII''''' | ||
| + | |||
| + | The high number of scaffolds is likely due to the size of the WBC protein family. | ||
| + | |||
| + | |||
| + | EST DR067855: 00004; 00495; 01882; 00012; 07909; 00779; 00714; 11598; 00355; 02244; 00809; 02234; 00126; 00350; 02006; 01110; 00282; 00742; 01812; 00045; 08837; 00332; 02508; 00913; 00507; 00118; 02687; 00560; 00214; 01191; 01030; 01994; 00365; 00781; 00630; 00040; 01209; 01838; 00881; 10517; 00227; 00262; 00504; 00927; 01770; 00038; 00368; 03502 | ||
| + | |||
| + | |||
| + | EST CV190423: 00560; 09668; 01965; 01191; 00913; 00896; 00263; 00227; 01994; 00040; 00365; 00087 | ||
| + | |||
| + | |||
| + | EST CF811331: 00779; 00809; 00355; 00714; 02006; 11598; 00004; 07909; 00495; 08837; 02234; 00012; 01882; 00045; 00126; 00350; 01110; 02244; 01812; 02508; 00282; 00742; 00332; 00913; 00507; 00118; 02687; 01030; 00560; 00214; 01191; 00365; 00781; 00630; 01838; 10517; 00227; 00881; 01209 | ||
| + | |||
| + | |||
| + | EST DR067166: 00087; 00040; 01994; 00365; 00227; 02687; 00504; 00560; 00118; 09668; 01965; 01030; 00131; 00507; 00116; 01016; 00555; 00913 | ||
| + | |||
| + | |||
| + | '''''CER6''''' | ||
| + | |||
| + | |||
| + | EST CF811643: 00137; 00431; 00133; 01322; 02309; 03134 | ||
| + | |||
| + | |||
| + | '''''MAH1''''' | ||
| + | |||
| + | |||
| + | EST DR066987: 00314; 01091; 04227; 09505; 00112; 00236; 00254; 00277 | ||
| + | |||
| + | |||
| + | EST CV090972: 00236; 00730; 01607; 00254 | ||
Latest revision as of 06:24, 17 April 2012
Blueberry coat color
Evidence presented by Albrigo et al. (Media:Albrigo_color.pdf) points to "waxy bloom" contributing highly to coat color in highbush blueberries. Other possible variables (such as pH and anthocyanins) were implicated in juice color but showed little to no influence on coat color. Also, a direct connection between beta-diketone presence and coat color was made by Sapers et al. (Media:Sapers_color.pdf). Berries with more beta-diketones displayed a higher degree of blue hue. Therefore, genes involved in wax production and secretion and beta-diketone synthesis were the focus of my research.
Samuels et al. (Media:Samuels_color.pdf) provided a thorough analysis of wax production in Arabidopsis. Additionally, pathways presented within the paper and the KEGG website indicated genes involved specifically in ketone production. Based on the relatedness of two genera, I searched for orthologs of Arabidopsis within the Vaccinium scaffold collection. Genes were chosen to represent a variety of roles within wax synthesis and secretion.
Target genes: WBCII (transporter: wax secretion); CER6 (fatty acid elongation); WSD (wax production); FATB (wax production); MAH1 (ketone production)
Source: Media:Samuels_color.pdf
Source: Media:Samuels_color.pdf
Source: KEGG
Finding orthologs
Samuels et al. provided MIPS accession numbers for each gene. Entering each number into The Arabidopsis Information Resource provides links to FASTA sequences for the gene in Arabidopsis. Scaffolds of the V. corymbosum genome containing possible orthologs were found using the tBLASTx tool on the Vaccinium website. Scaffolds were chosen based on E-score. SSRs (and primers for each SSR) were found for each scaffold using the SSR tool on the Vaccinium website. SSRs were chosen based on proximity to the region of the scaffold aligning with the submitted Arabidopsis gene. Also, di- and trinucleotide repeats of 5 or greater were favored and the size of PCR products was required to fall between 100 and 700 bp.
Primers
WBCII
Scaffold 00913 (Aligning region: 72307 - 93373 bp)
1. Forward Primer GTGCTAGAATCCCCTGAAACAC & Reverse Primer CCCCCTCTCTCTTTCTTCCTAT (ga) x5 PCR product = 165 (64928 - 64937 bp)
2. Forward Primer GTGTGCATCTCTCTCTGATTGC & Reverse Primer TCTCTCCCATCTTAAAACCCAA (ga) x5 PCR product = 132 (96985 - 96994 bp)
3. Forward Primer CCGTAAGTAGCGAGGAGACTGT & Reverse Primer ACAAATTCGGACGGAGAGAGTA (ga) x9 PCR product = 227 (100495 - 100512 bp)
Scaffold 01191 (Aligning region: 8751 - 13731 bp)
1. Forward Primer TCTTATTGGTCCTCGGTGCTAT & Reverse Primer CACAAAATGGCCTAAGTAGGGA (ca) x5 PCR product = 258 (3896 - 3905 bp)
2. Forward Primer ACTGGAAATGGTAGAAATGGGA & Reverse Primer GAGGAAGAGGAAAAAGGAGGAG (ga) x15 PCR product = 284 (14258 - 14287 bp)
CER6
Scaffold 00431 (Aligning region: 45978 - 48317 bp)
1. Forward Primer ATCCGACTTGGTAATTTGATGG & Reverse primer CGGAATGAACACCAACAATAGA (ct) x8 PCR product = 257 (36598 - 36613 bp)
2. Forward Primer TGCTGAGTGGTGGAGTTCATTA & Reverse primer AAGCCTTACCGCTCTCCTAACT (att) x5 PCR product = 267 (55128 - 55142 bp)
Scaffold 00268 (Aligning region: 190730 - 192192 bp)
1. Forward Primer ATTGCAGATCCTTCCAGTCAAT & Reverse primer TTAACATGCCATTGTCGAAGAC (tg) x5 PCR product = 222 (179241 - 179250 bp)
2. Forward Primer TTTGACGTACTTGAGGTTCACG & Reverse primer AAAGAGAGGAGAGAGAGGGAGG (tc) x5 PCR product = 292 (192395 - 192404 bp)
Scaffold 00137 (Aligning region: 145956 - 147476 bp)
1. Forward Primer AACACCTTGACGAACTGAACCT & Reverse primer CAATCCAGTATTGAGACAGCCA (tc) x5 PCR product = 107 (143881 - 143890 bp)
2. Forward Primer AAATTGCTAGAGTTGCCGTTGT & Reverse primer ATCACATGGACTGGTAGAGCCT (tg) x5 PCR product = 139 (158644 - 158653 bp)
WSD
Scaffold 00550 (Aligning region: 130667 - 130867 bp)
1. Forward Primer ATGTTATTCCATCCGCCTCTAA & Reverse primer ACCTCAAACAAATAACCCAACG (ga) x5 PCR product = 190 (129038 - 129047 bp)
2. Forward Primer AATTGAGAAATGTGAGTCCCGT & Reverse primer GAGACACGCTAAGATTTGCCTT (at) x5 PCR product = 295 (135042 - 135051 bp)
FATB
Scaffold 00698 (Aligning region: 13633 - 18095 bp)
1. Forward Primer GAAGTCTTCACCACTCCGATTC & Reverse Primer TACCACGCAAAATTATCACCAG (tc) x5 PCR product = 281 (11438 - 11447 bp)
2. Forward Primer TCCCAATCCAATTGATATCCTC & Reverse Primer TCACTCACCTCTTCCAAGGATT (at) x7 PCR product = 259 (19774 - 19787 bp)
MAH1
Scaffold 01091 (Aligning region: 124548 - 125522 bp)
1. Forward Primer TGTATTAAAGTGTGTGGGGGTG & Reverse Primer TAGAACACGAAAATGGTGTTGG (tc) x11 PCR product = 184 (95581 - 95602 bp)
2. Forward Primer ATAGGCAAAGAATTTTGGGAGG & Reverse Primer TACACACACACACACACACACA (gtat) x8 PCR product = 129 (130046 - 130077 bp)
Scaffold 00236 (Aligning region: 226568 - 227989 bp)
1. Forward Primer ACCACCACCACCTCTCTCTCT & Reverse Primer AAGGTTTTGATGGCTTACCTCA (tca) x7 PCR product = 148 (226187 - 226207 bp)
2. Forward Primer CTCTTCTCCCCCTTTCATACCT & Reverse Primer CTTCCTGGTGAAGAGAAATTGG (ta) x5 PCR product = 225 (230813 - 230822 bp)
ESTs
Expressed sequence tags (ESTs) were found using tBLASTn against the EST database of the Vaccinium website. Queries were generated by copying the database sequence from a section of amino acids aligning with the target gene. Confirmation of any EST hits with a sufficient E-score was made using the Unigene search feature of the Vaccinium website.
No ESTs were found for WSD or FATB.
WBCII
Scaffold 01191: DR067855
Scaffold 01191: CV190423
Scaffold 01191: CF811331
Scaffold 00913: DR067166
CER6
Scaffold 00137 and 00268: CF811643
MAH1
(Cytochrome P450 listed as alternate name for MAH1 on The Arabidopsis Information Resource)
Scaffold 01091: DR066987
Scaffold 00236: CV090972
Scaffolds found using ESTs
The following represent scaffolds that may indicate paralogs of each gene within the blueberry genome. They were found by submitting the FASTA of the EST into a tBLASTx against the Vaccinium scaffold database.
WBCII
The high number of scaffolds is likely due to the size of the WBC protein family.
EST DR067855: 00004; 00495; 01882; 00012; 07909; 00779; 00714; 11598; 00355; 02244; 00809; 02234; 00126; 00350; 02006; 01110; 00282; 00742; 01812; 00045; 08837; 00332; 02508; 00913; 00507; 00118; 02687; 00560; 00214; 01191; 01030; 01994; 00365; 00781; 00630; 00040; 01209; 01838; 00881; 10517; 00227; 00262; 00504; 00927; 01770; 00038; 00368; 03502
EST CV190423: 00560; 09668; 01965; 01191; 00913; 00896; 00263; 00227; 01994; 00040; 00365; 00087
EST CF811331: 00779; 00809; 00355; 00714; 02006; 11598; 00004; 07909; 00495; 08837; 02234; 00012; 01882; 00045; 00126; 00350; 01110; 02244; 01812; 02508; 00282; 00742; 00332; 00913; 00507; 00118; 02687; 01030; 00560; 00214; 01191; 00365; 00781; 00630; 01838; 10517; 00227; 00881; 01209
EST DR067166: 00087; 00040; 01994; 00365; 00227; 02687; 00504; 00560; 00118; 09668; 01965; 01030; 00131; 00507; 00116; 01016; 00555; 00913
CER6
EST CF811643: 00137; 00431; 00133; 01322; 02309; 03134
MAH1
EST DR066987: 00314; 01091; 04227; 09505; 00112; 00236; 00254; 00277
EST CV090972: 00236; 00730; 01607; 00254