Difference between revisions of "BioBrick Digestions for Fragment and Vector Preparation"
From GcatWiki
| Line 5: | Line 5: | ||
# Part B in vector cut with SP, followed by part A as insert cut with XP | # Part B in vector cut with SP, followed by part A as insert cut with XP | ||
# Part B as insert cut with RS, followed by part A in vector cut with RX | # Part B as insert cut with RS, followed by part A in vector cut with RX | ||
| + | |||
| + | [[Image: BioBrick_assembly.JPG]] | ||
We use enzymes from [http://www.neb.com/nebecomm/products/category1.asp?#150 New England Biolabs] that are High Fidelity. Each of the four enzymes cuts in the same buffer, NEB 4. | We use enzymes from [http://www.neb.com/nebecomm/products/category1.asp?#150 New England Biolabs] that are High Fidelity. Each of the four enzymes cuts in the same buffer, NEB 4. | ||
Latest revision as of 17:32, 12 July 2010
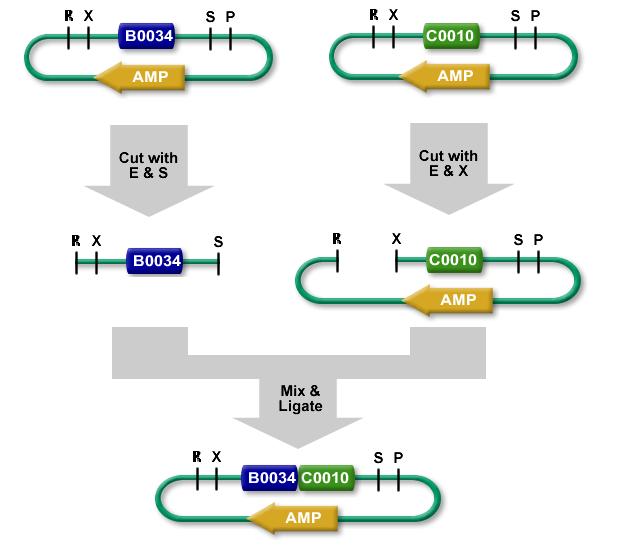
There are four ways to put two BioBrick parts together with the most commonly used Assembly Standard. The enzymes EcoRI (R), XbaI (X), SpeI (S), and PstI (P) can be used for these assemblies.
- Part A in vector cut with SP, followed by part B as insert cut with XP
- Part A as insert cut with RS, followed by part B in vector cut with RX
- Part B in vector cut with SP, followed by part A as insert cut with XP
- Part B as insert cut with RS, followed by part A in vector cut with RX
We use enzymes from New England Biolabs that are High Fidelity. Each of the four enzymes cuts in the same buffer, NEB 4.
Use the following procedure for a digestion.
- Transfer about 2000 ng into a 1.5 ml tube and make up the difference with pure dH2O to 30 ul, then add the following:
4 ul NEB4 10X buffer 4 ul 10X BSA 1 ul HF first enzyme 1 ul HF second enzyme
- Incubate at 37 C for 30 minutes
- Add 5 ul blue sample buffer to each and run on gel