Difference between revisions of "Can we solve a 3-SAT problem with supressor logic?"
(→How did Sakamoto et al. use a DNA computer to solve a 3-SAT problem?) |
(→How did Sakamoto et al. use a DNA computer to solve a 3-SAT problem?) |
||
| Line 6: | Line 6: | ||
| − | + | <center> [[Image:hairpin.GIF]]<br></center> | |
Sakamoto used hairpin formations in single stranded DNA (ssDNA) as a molecular computer. Hairpin structures are formed when complimentary bases on the same strand attach to each other forming a loop. As shown in Picture B above, C and -C are compliments of each other, and bind together. Picture A is what a normal ssDNA should look like. Sakamoto used this self assembly of secondary structures on a satisfiability (SAT) problem. The problem had six inputs and ten clauses. An example of a clause would be (a or b or -c), and could include any combinations of inputs from a to f including -a to -f. If the problem is satisfied, the ssDNA stays in it's normal form. If the problem is not satisfied, the ssDNA forms a hairpin. Later in the paper it explains that the hairpin forming molecules can be removed from the others with certain techniques. | Sakamoto used hairpin formations in single stranded DNA (ssDNA) as a molecular computer. Hairpin structures are formed when complimentary bases on the same strand attach to each other forming a loop. As shown in Picture B above, C and -C are compliments of each other, and bind together. Picture A is what a normal ssDNA should look like. Sakamoto used this self assembly of secondary structures on a satisfiability (SAT) problem. The problem had six inputs and ten clauses. An example of a clause would be (a or b or -c), and could include any combinations of inputs from a to f including -a to -f. If the problem is satisfied, the ssDNA stays in it's normal form. If the problem is not satisfied, the ssDNA forms a hairpin. Later in the paper it explains that the hairpin forming molecules can be removed from the others with certain techniques. | ||
Revision as of 20:35, 7 April 2009
Contents
What is the 3-SAT problem?
How did Sakamoto et al. use a DNA computer to solve a 3-SAT problem?
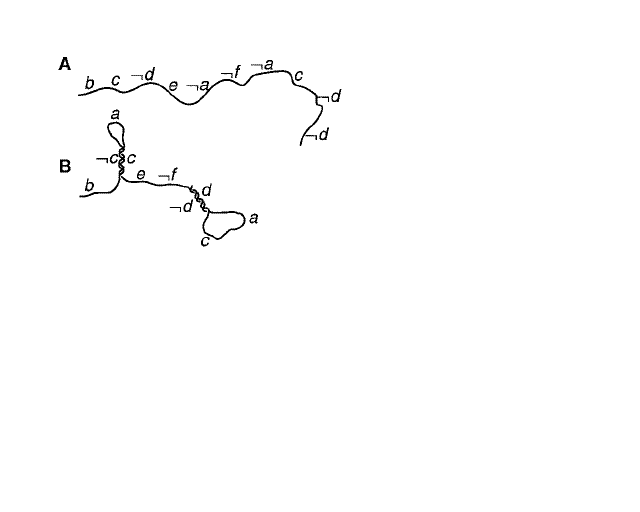
Sakamoto used hairpin formations in single stranded DNA (ssDNA) as a molecular computer. Hairpin structures are formed when complimentary bases on the same strand attach to each other forming a loop. As shown in Picture B above, C and -C are compliments of each other, and bind together. Picture A is what a normal ssDNA should look like. Sakamoto used this self assembly of secondary structures on a satisfiability (SAT) problem. The problem had six inputs and ten clauses. An example of a clause would be (a or b or -c), and could include any combinations of inputs from a to f including -a to -f. If the problem is satisfied, the ssDNA stays in it's normal form. If the problem is not satisfied, the ssDNA forms a hairpin. Later in the paper it explains that the hairpin forming molecules can be removed from the others with certain techniques.
What is suppressor logic?
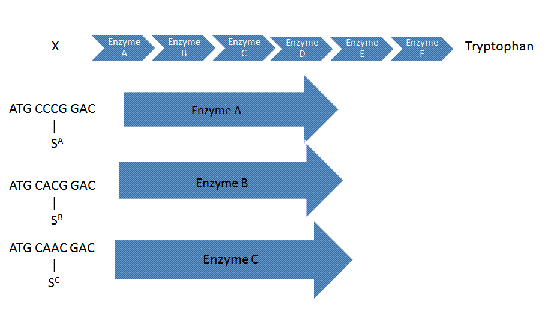
Suppressor Suppressor logic uses suppressor tRNAs to avoid frameshift mutations in an amino acid sequence.
A frameshift is a genetic mutation caused by the addition or deletion of nucelotides to a given sequence which codes for a protein. Since codons are read in a series of three, the addition or deletion of nucleotides will disrupt the reading frame. This disruption will most likely cause the production of a nonfunctional protein.

A frameshift occurs and, in this case, a guanine is added to the sequence. If nothing is done, enzyme A will not be made, meaning the clause will not be satisfied.
The suppressor tRNA allows the 4 letter sequence to be read as a single codon, therefore, keeping the protein on track.
How could suppressor logic be used to solve the Sakamoto 3-SAT problem?
Definitions
Inputs = framshift suppressor tRNAs
Input value = supp a is 1, supp g is 0; supp b is 1, supp h is 0, etc. up to tth 6th pair of f and l
Logical clause (LC) = three inputs connected by OR, eg. (a OR b OR e)
Logical expression (LE) = string of LCs connected by AND
Subroutine
1. Individual bacteral cells use Hin/hix system to randomly choose of of the 64 possible combinations of 6 inputs.
2. Each bacterial cell carries out the following subroutine on each LC: IF LC=TRUE THEN "check the next LC" ELSEIF LC=FALSE "go get a new set of inputs with step 1"
3. If/when a bacterial cell finds a set of inputs that satisfies the entire LE (ie. a solution to the 3-SAT problem), it will glow green.