Difference between revisions of "Chitinase"
From GcatWiki
| Line 2: | Line 2: | ||
[[Image:pathway.png]]<br> | [[Image:pathway.png]]<br> | ||
The pathway above is labeled for a halophile, haloarcula marismortui (most of the other halophiles on KEGG did not have a reference pathway for Aminosugars Metabolism). As you can see, Chitinase (EC#:3.2.1.14) is not predicted for this halophile. | The pathway above is labeled for a halophile, haloarcula marismortui (most of the other halophiles on KEGG did not have a reference pathway for Aminosugars Metabolism). As you can see, Chitinase (EC#:3.2.1.14) is not predicted for this halophile. | ||
| + | <br> | ||
| + | The only automated annotation database to predict a Chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810.<br> | ||
| + | The top BlastP results: [[Image:blastpr.jng]] | ||
Revision as of 15:47, 6 November 2008
KEGG Pathway containing Chitinase: Aminosugars Metabolism
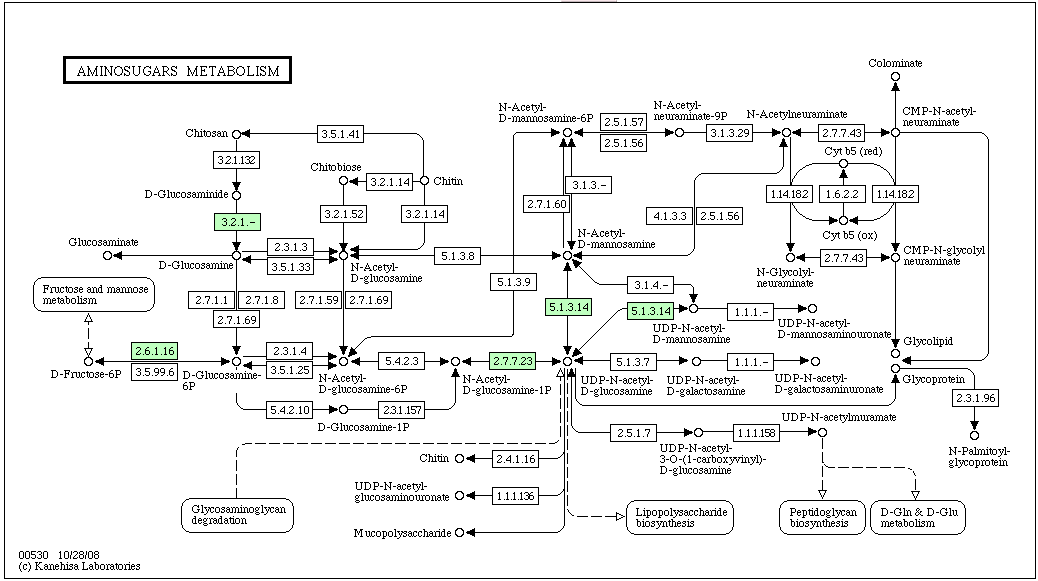
The pathway above is labeled for a halophile, haloarcula marismortui (most of the other halophiles on KEGG did not have a reference pathway for Aminosugars Metabolism). As you can see, Chitinase (EC#:3.2.1.14) is not predicted for this halophile.
The only automated annotation database to predict a Chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810.
The top BlastP results: File:Blastpr.jng