Difference between revisions of "Chitinase"
| Line 9: | Line 9: | ||
So, although it is possible that Chitinase is coded for by this region, there is a stronger possibility that it is not.<br> | So, although it is possible that Chitinase is coded for by this region, there is a stronger possibility that it is not.<br> | ||
<br><br> | <br><br> | ||
| − | While doing some hand-curated annotations, we also found another potential Chitinase-coding region. | + | While doing some hand-curated annotations, we also found another potential Chitinase-coding region at 72986...74821 (+). |
| + | [[Image:ChitinaseBlast]] | ||
Revision as of 15:49, 18 November 2008
KEGG Pathway containing Chitinase: Aminosugars Metabolism
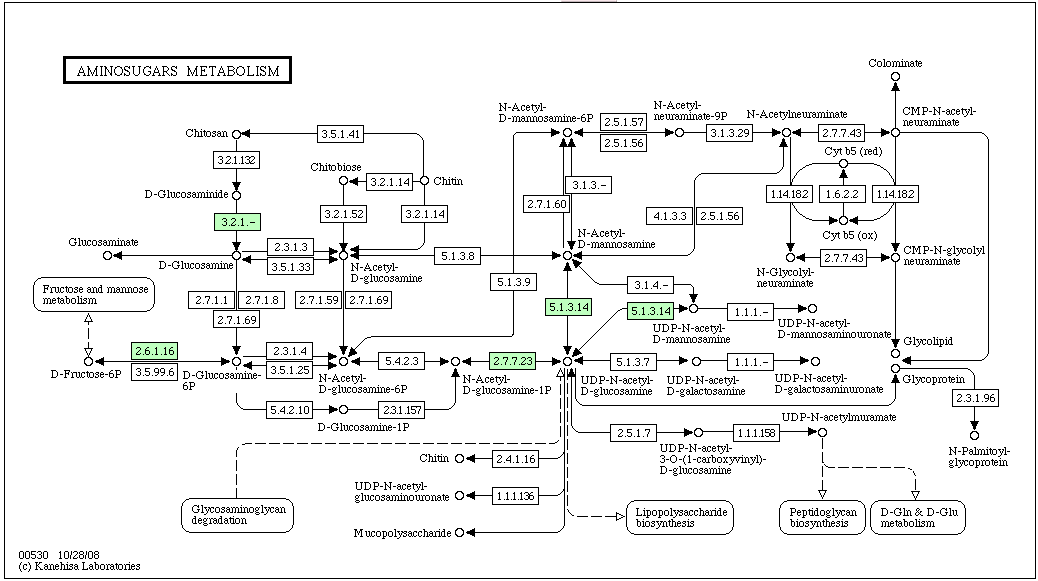
The pathway above is labeled for a halophile, haloarcula marismortui (most of the other halophiles on KEGG did not have a reference pathway for Aminosugars Metabolism). As you can see, Chitinase (EC#:3.2.1.14) is not predicted for this halophile.
The only automated annotation database to predict a Chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810. Chitinase A: Manatee
The top BlastP results: 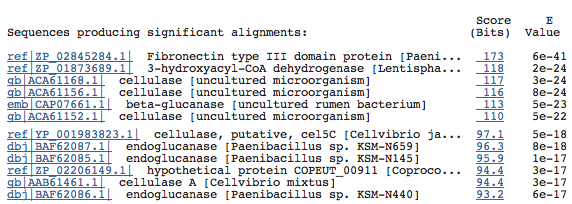
As you can see, Chitinase A is not among the top hits. However, there is one hit for it further down the list:
![]()
So, although it is possible that Chitinase is coded for by this region, there is a stronger possibility that it is not.
While doing some hand-curated annotations, we also found another potential Chitinase-coding region at 72986...74821 (+).
File:ChitinaseBlast