Difference between revisions of "Chitinase"
| Line 5: | Line 5: | ||
The only automated annotation database to predict a chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810. [http://www.tigr.org/tigr-scripts/prok_manatee/shared/ORF_infopage.cgi?db=nthu01&orf=ORF00052 Chitinase A: Manatee]<br> | The only automated annotation database to predict a chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810. [http://www.tigr.org/tigr-scripts/prok_manatee/shared/ORF_infopage.cgi?db=nthu01&orf=ORF00052 Chitinase A: Manatee]<br> | ||
The top BlastP results: [[Image:blastpr.png]]<br><br> | The top BlastP results: [[Image:blastpr.png]]<br><br> | ||
| − | As you can see, Chitinase A is not among the top hits. However, there is one hit for it further down the list:<br> | + | As you can see, Chitinase A is not among the top hits. However, there is one hit for it further down the list with a good e-value:<br> |
[[Image:blastpr2.png]]<br> | [[Image:blastpr2.png]]<br> | ||
So, this location possibly codes for chitinase.<br> | So, this location possibly codes for chitinase.<br> | ||
| Line 13: | Line 13: | ||
Second Chitinase hit: [[Image:ChitinaseBlast2.png]] <br> <br> | Second Chitinase hit: [[Image:ChitinaseBlast2.png]] <br> <br> | ||
| − | + | Both of these hits had extremely good e-values; so, data from this coding region strongly suggest the coding of chitinase in our organism. However, we do not yet have any physiological data to support the presence of chitinase. | |
Latest revision as of 15:13, 4 December 2008
KEGG Pathway containing Chitinase: Aminosugars Metabolism
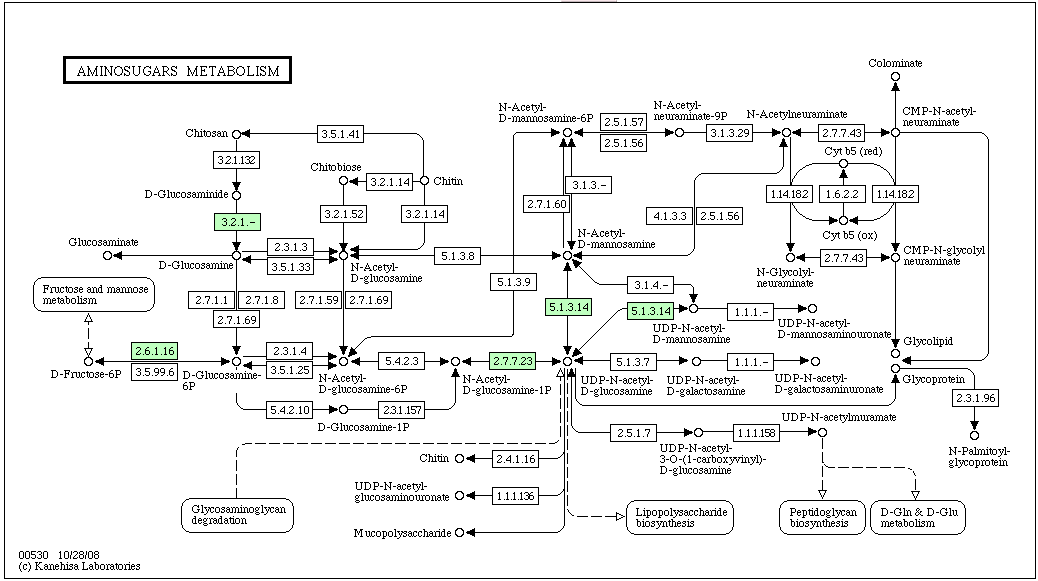
The pathway above is labeled for a halophile, haloarcula marismortui (most of the other halophiles on KEGG did not have a reference pathway for Aminosugars Metabolism). As you can see, chitinase (EC#:3.2.1.14) is not predicted for this halophile.
The only automated annotation database to predict a chitinase was Manatee. Manatee predicted Chitinase A at 55855...57810. Chitinase A: Manatee
The top BlastP results: 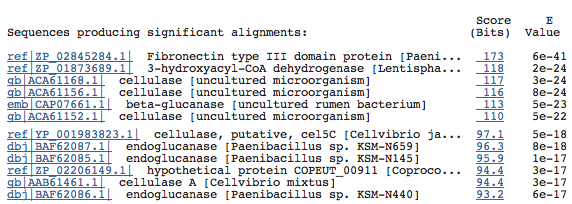
As you can see, Chitinase A is not among the top hits. However, there is one hit for it further down the list with a good e-value:
![]()
So, this location possibly codes for chitinase.
While doing some hand-curated annotations, we also found another potential chitinase-coding region at 72986...74821 (+). Two hits were found by Blastp that coded for chitinase, they were the 2nd and 3rd hits overall and both were from halophiles:
First Chitinase hit: 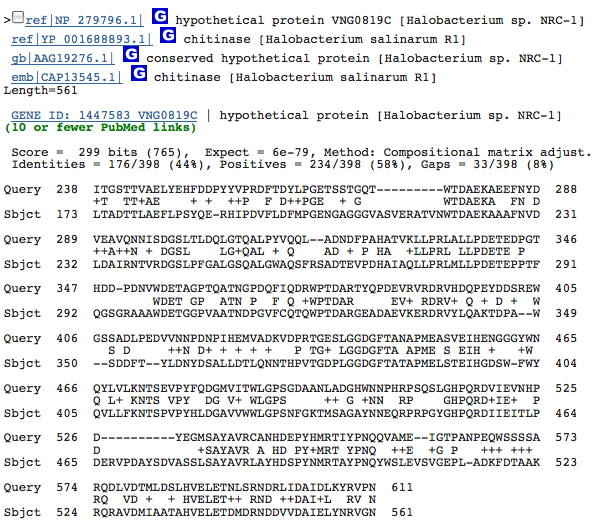
Second Chitinase hit: 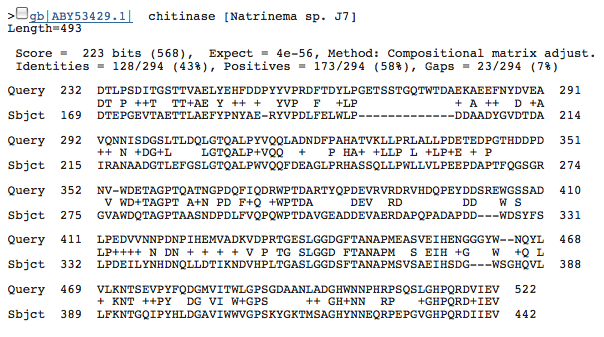
Both of these hits had extremely good e-values; so, data from this coding region strongly suggest the coding of chitinase in our organism. However, we do not yet have any physiological data to support the presence of chitinase.