Difference between revisions of "Claudia's Assignment"
Clcarcelen (talk | contribs) (→Introduction) |
Clcarcelen (talk | contribs) |
||
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
| − | From the background information on our species, ''Halomicrobium mukohataei'', we know that our organism is not only a halophile, or salt-loving, but also that the organism in fact requires a high salinity environment to survive. This trait is unique and compelling because of the harsh environment and the many pumps and mechanisms that must be present for an living organism to survive. However, our organism is not the only one to have such adaptations. There are a number of halophile species as well as a number of salt-tolerant organisms, including bacteria and yeasts. | + | From the background information on our species, ''Halomicrobium mukohataei'', we know that our organism is not only a halophile, or salt-loving, but also that the organism in fact requires a high salinity environment to survive. This trait is unique and compelling because of the harsh environment and the many pumps and mechanisms that must be present for an living organism to survive. However, our organism is not the only one to have such adaptations. There are a number of halophile species as well as a number of salt-tolerant organisms, including a number of bacteria and a few yeasts.<br> |
| + | Originally, I wanted to look into the genes that encoded adaptations to the high-salinity environment, specifically ion (cation) pumps. Of the proteins with known functions, our organism appears to encode 22 different proteins functioning as ion or ion-dependent pumps. I wanted to investigate these pumps, and their roles within the organism's metabolic pathways. | ||
==Cation Pumps== | ==Cation Pumps== | ||
Revision as of 01:47, 1 October 2009
Introduction
From the background information on our species, Halomicrobium mukohataei, we know that our organism is not only a halophile, or salt-loving, but also that the organism in fact requires a high salinity environment to survive. This trait is unique and compelling because of the harsh environment and the many pumps and mechanisms that must be present for an living organism to survive. However, our organism is not the only one to have such adaptations. There are a number of halophile species as well as a number of salt-tolerant organisms, including a number of bacteria and a few yeasts.
Originally, I wanted to look into the genes that encoded adaptations to the high-salinity environment, specifically ion (cation) pumps. Of the proteins with known functions, our organism appears to encode 22 different proteins functioning as ion or ion-dependent pumps. I wanted to investigate these pumps, and their roles within the organism's metabolic pathways.
Cation Pumps
The following is a list of the cation pumps and cation-influenced structures encoded in our organism's genome:
Na+/proline symporter 644032116
A BLAST (megablast) of the gene encoding this ion pump produced the following results:

Kef-type K+ ransport system, predicted NAD-binding component 644032129
A BLAST using megablast produced no results other than our own species.
A BLAST (blastn) of the gene encoding this ion pump produced the following results:
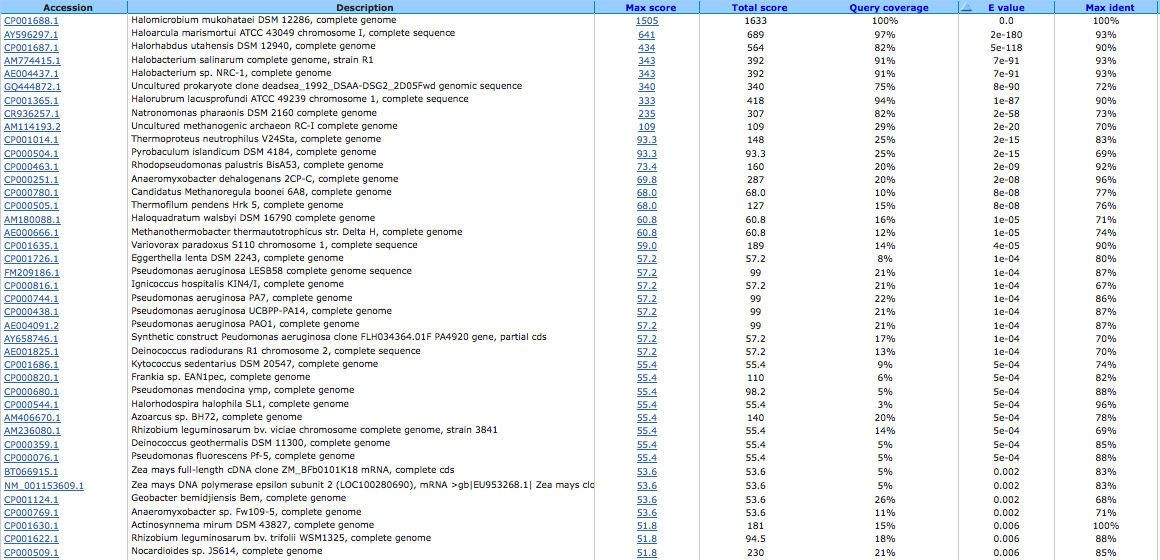
Ca2+/Na+ antiporter 644032240
A BLAST using megablast produced no results other than our own species.
A BLAST using blastn produced the following results:
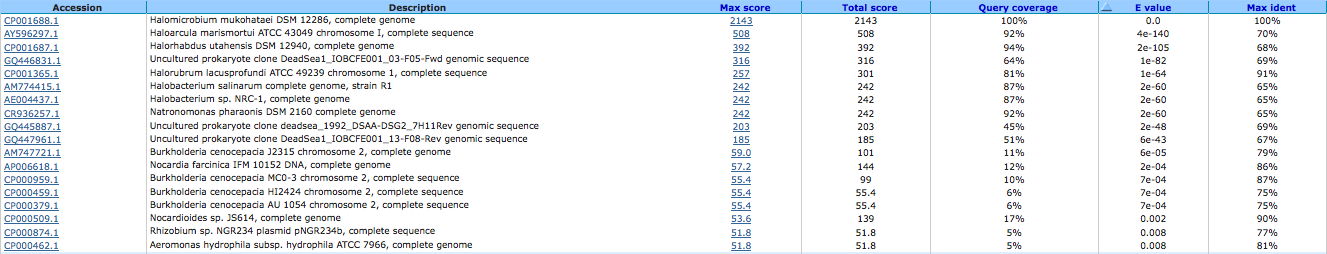
Mn2+/Fe2_ transporter, NRAMP family 644032307
A BLAST using megablast produced the following results:
![]()
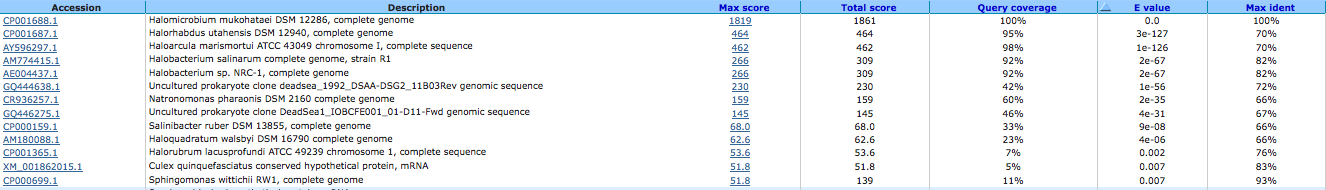
H(+)-transporting ATP synthase, vacuolar type, subunit D ( EC:3.6.3.14 ) 644032373
A BLAST using megablast produced the following results:

A BLAST using blastn produced the following results:
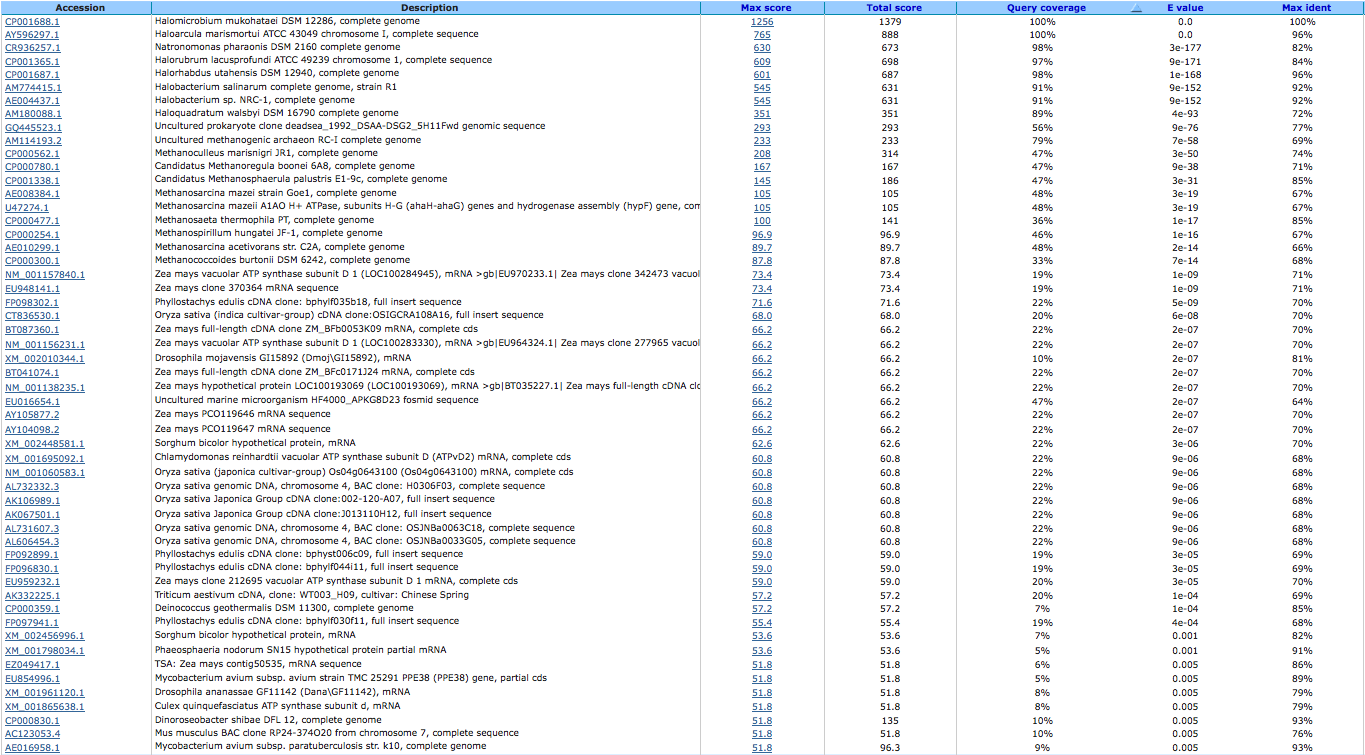
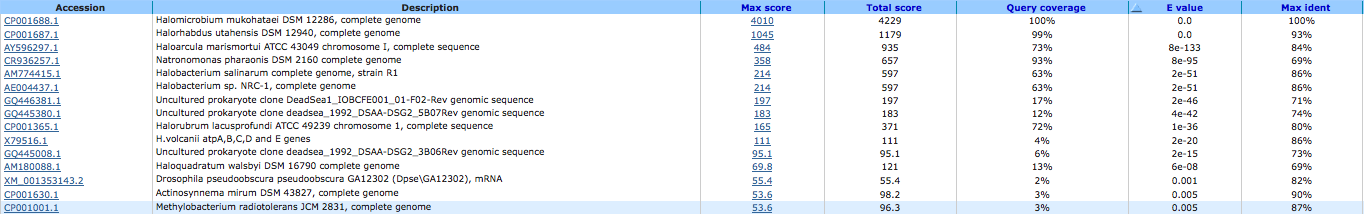
archaeal/vacuolar-type H+-ATPase subunit B ( EC:3.6.3.14 ) 644032378
A BLAST using megablast produced the following results:

A BLAST using blastn produced the following results:
archaeal/vacuolar-type H+-ATPase subunit F ( EC:3.6.3.14 ) 644032380
A BLAST using megablast produced no results other than our own species.
A BLAST using blastn produced the following results:
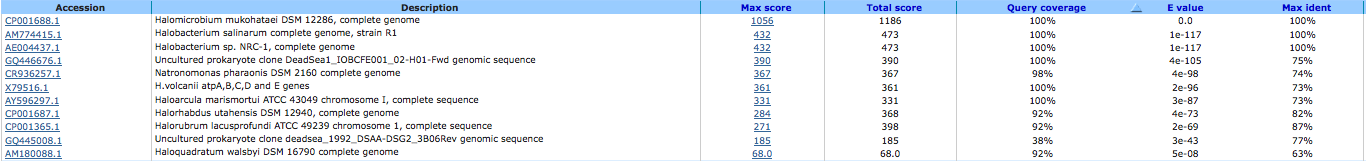
archaeal/vacuolar-type H+-ATPase subunit E ( EC:3.6.3.14 ) 644032382
A BLAST using megablast produced no results other than our own species.
A BLAST using blastn produced the following results:
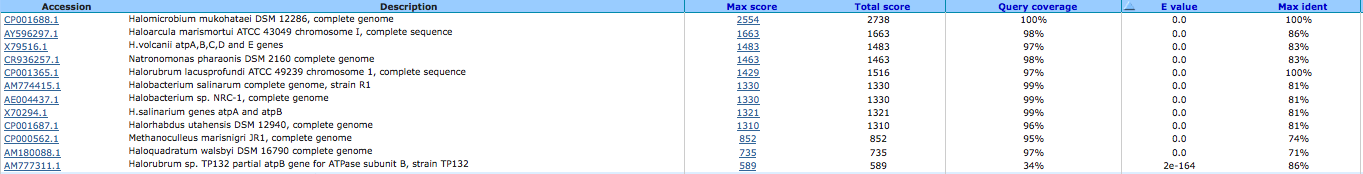
archaeal/vacuolar-type H+-ATPase subunit I ( EC:3.6.3.14 ) 644032384
A BLAST using megablast produced the following results:

A BLAST using blastn produced the following results:
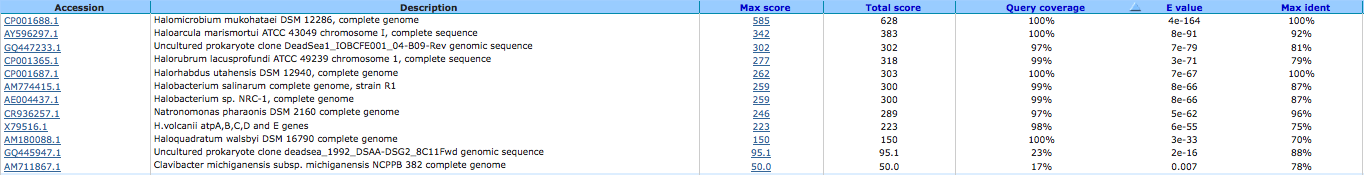
K+ transport system, NAD-binding component 644032472
A BLAST using megablast produced no results other than our own species.
A BLAST using blast n produced the following results:
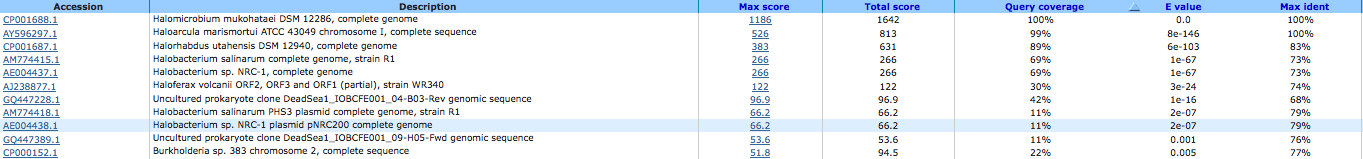
NAD+ synthetase ( EC:6.3.5.1 ) 644032488
A BLAST using megablast produced no results other than our own species.
A BLAST using blastn produced the following results:
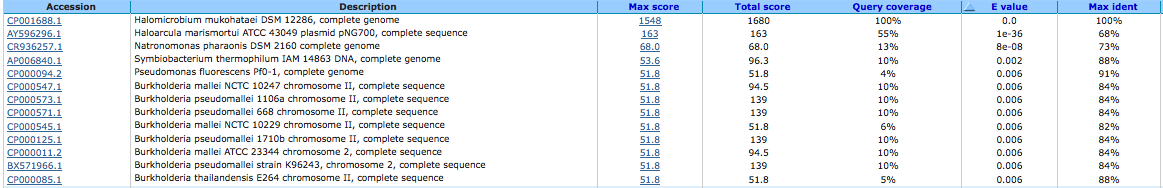
NH(3)-dependent NAD(+) synthetase (EC 6.3.1.5) (IMGterm) 644032494
A BLAST using megablast produced the following results:


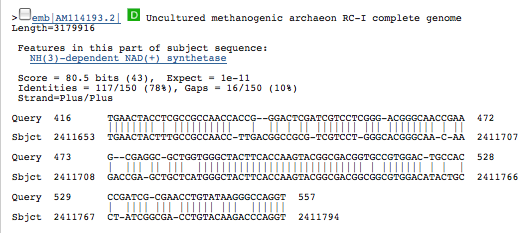
A BLAST using blastn produced the following results: