Davidson Projects with Updates
Contents
- 1 Building LuxI sender TESTER cell
- 2 Building LuxR receiver TESTER cell
- 3 Building LuxR constitutive protein
- 4 Building LasI sender TESTER cell
- 5 Building LasR receiver TESTER cell
- 6 Building LasR constitutive protein
- 7 Building LsrK/LsrR receiver TESTER cell
- 8 Testing Crosstalk between Lux and Las
- 9 AND gate using pTetLac promoter
- 10 Building XOR Gate
- 11 Testing XOR Gate
- 12 Making Better pLac promoter and LacI proteins
Building LuxI sender TESTER cell
| Part # | Colonies from Registry Recovery? | Gel Verification? | Status of Testing the Device | Experimental Outcome |
|---|---|---|---|---|
| S03608 | Yes | Yes | Ready | Successful |
Building LuxR receiver TESTER cell
| Colonies from Registry Recovery? | Gel Verification? | Status of Testing the Device | Experimental Outcome | |
|---|---|---|---|---|
| K09100 | Yes | Yes | Ready | Successful |
Building LuxR constitutive protein
Promoter (pBad?) + Fix this
| Colonies from Registry Recovery? | Gel Verification? | Status of Testing the Device | Experimental Outcome | |
|---|---|---|---|---|
| Fix This | ? | ? | ? | ? |
| Fix This | ? | ? | ? | ? |
Building LasI sender TESTER cell
R0011 + B0034 + C0178.
File:InsertHere.jpg
| Colonies from Registry Recovery? | Gel Verification? | Successful Ligation? | Status of Testing the Device | |
|---|---|---|---|---|
| R0011 | Yes | Yes | ||
| B0034 | Yes | Yes | ||
| C0178 | Yes | Yes |
Building LasR receiver TESTER cell
| Colonies from Registry Recovery? | Gel Verification? | Successful Ligation? | Status of Testing the Device | |
|---|---|---|---|---|
| S03156 | Yes | Yes | Yes | |
| B0015 | Yes | Yes | Yes | |
| R0079 | Yes | Yes | Yes | |
| E0240 | Yes | Yes | Yes | |
| R0011 | Yes | Yes | Pending |
Ligation 1A S03156 + B0015
Ligation 1B R0079 + E0240
Ligation 1C B0015 + R0079
Done!
A
![]()
B
![]()
C
![]()
Ligation 2: Successful Ligation 1A + Successful Ligation 1B
Done!
![]()
Ligation 3: R0011 + Ligation 2
Building LasR constitutive protein
Promoter J23100
| Colonies from Registry Recovery? | Gel Verification? | Status of Testing the Device | Experimental Outcome | |
|---|---|---|---|---|
| J23100 | Yes | Yes | ? | ? |
| S03954 | Yes | Yes | Ready | Pending |
Building LsrK/LsrR receiver TESTER cell
LsrKp + RBS + LsrK + TT
LsrRp + RBS + LsrR + reporter (GFP/RFP/YFP) + TT
LsrK needs to be constitutively expressed in order to receive the AI-2 signal. LsrR acts as its own repressor, so the reporter should not be activated until the AI-2 signal is received. Once LsrR binds to AI-2 molecule, the LsrR gene is activated and the positive feedback loop is initiated, which also turns on the reporter gene.
| Colonies from Registry Recovery? | Gel Verification? | Status of Testing the Device | Experimental Outcome | |
|---|---|---|---|---|
| part no. | ? | ? | ? | ? |
Testing Crosstalk between Lux and Las
1) Mix LuxI sender cells with LasR receiver tester cells: negative control
2) Mix LasI sender cells with LuxR receiver tester cells: negative control
3) Mix LuxI sender cells with LuxR receiver tester cells: experiment we hope will glow green
4) Mix LasI sender cells with LasR receiver tester cells: experiment we hope will glow green
5) Verify LuxR receiver tester cells don't glow solo
6) Verify LasR receiver tester cells don't glow solo
AND gate using pTetLac promoter
We have successfully built the pTetLac AND promoter: K091101.
This is sequence verified.
Now we need the various LacI proteins (I12, X86 and I12+X86 variants).
We need tetR repressor protein. We have tried to get it from the paper registry.
We need to see if this AND gate works as designed.
We need to test output to see if the two halves are balanced.
Building XOR Gate
List of auto-inducers and their catalog numbers.
Here is an idea Malcolm and Laurie developed.
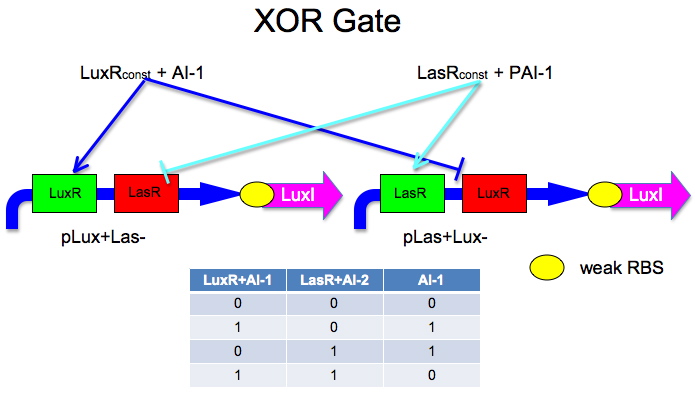
The idea is to have two mirrored halves of the system. LasR is regulated by PAI-1 {3-oxododecanoyl-HSL (3OC12HSL)} and LuxR is activated by AI-1 {3-oxohexanoyl-homoserine lactone (3OC6HSL)}. There is a potential problem in that the Lux half is more likely to get positive feedback than the Las half. This MAY not be a problem because 0/0 is leaky so we put a weak RBS to minimize leaky protein production. Also, if we add AI-2 and AI-1 is produced by leak, then the entire system shuts down. The repressor site is located between -35 and -10 of the promoter. The activator binding site is upstream of -35. This has been documented by Egland and Greenberg
Sequences we will need to make this XOR gate.
| Part | Oligos | Oligo Design |
|---|---|---|
| lpLas' | 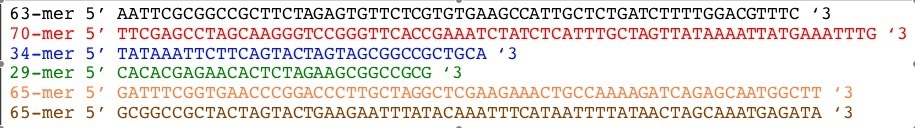 |
|
| pLasXOR | 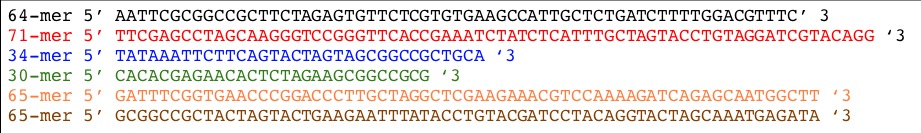 |
|
| Part | Forward Primer | Reverse Primer |
|---|---|---|
| pLux | 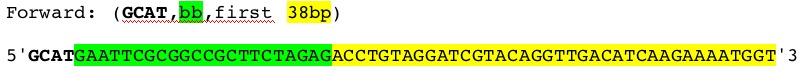 |

|
| pLuxXOR |  |

|
Testing XOR Gate
We will need to have constitutive LasR and LuxR made in these cells.
Then we will need to hit them with the two chemicals AHL types and commercial sources.
Making Better pLac promoter and LacI proteins
LacI wild-type gene sequence (ORF begins at the first amino acid)
ATGAAACCAGTAACGTTATACGATGTCGCAGAGTATGCCGGTGTCTCTTATCAGACCGTTTCCCGCGTGGTGAACCAGGCCAGCCACGTTTCTGCGAAAACGCGGGAAAAAGTGGAAGC
GGCGATGGCGGAGCTGAATTACATTCCCAACCGCGTGGCACAACAACTGGCGGGCAAACAGTCGTTGCTGATTGGCGTTGCCACCTCCAGTCTGGCCCTGCACGCGCCGTCGCAA
ATTGTCGCGGCGATTAAATCTCGCGCCGATCAACTGGGTGCCAGCGTGGTGGTGTCGATGGTAGAACGAAGCGGCGTCGAAGCCTGTAAAGCGGCGGTGCACAATCTTCTCGCGC
AACGCGTCAGTGGGCTGATCATTAACTATCCGCTGGATGACCAGGATGCCATTGCTGTGGAAGCTGCCTGCACTAATGTTCCGGCGTTATTTCTTGATGTCTCTGACCAGACACCATCA
ACAGTATTATTTTCTCCCATGAAGACGGTACGCGACTGGGCGTGGAGCATCTGGTCGCATTGGGTCACCAGCAAATCGCGCTGTTAGCGGGCCCATTAAGTTCTGTCTCGGCG
CGTCTGCGTCTGGCTGGCTGGCATAAATATCTCACTCGCAATCAAATTCAGCCGATAGCGGAACGGGAAGGCGACTGGAGTGCCATGTCCGGTTTTCAACAAACCATGCAAATGC
TGAATGAGGGCATCGTTCCCACTGCGATGCTGGTTGCCAACGATCAGATGGCGCTGGGCGCAATGCGCGCCATTACCGAGTCCGGGCTGCGCGTTGGTGCGGATATCTCGGTAGT
GGGATACGACGATACCGAAGACAGCTCATGTTATATCCCGCCGTTAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACCGCTTGCTGCAACTCTCTCAG
GGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCTGGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATGC
AGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGA
LacI_I12 Mutation (amino acid 3 is changed from CCA to TAT)
ATGAAATATGTAACGTTATACGATGTCGCAGAGTATGCCGGTGTCTCTTATCAGACCGTTTCCCGCGTGGTGAACCAGGCCAGCCACGTTTCTGCGAAAACGCGGGAAAAAGTGGAAGC
GGCGATGGCGGAGCTGAATTACATTCCCAACCGCGTGGCACAACAACTGGCGGGCAAACAGTCGTTGCTGATTGGCGTTGCCACCTCCAGTCTGGCCCTGCACGCGCCGTCGCAA
ATTGTCGCGGCGATTAAATCTCGCGCCGATCAACTGGGTGCCAGCGTGGTGGTGTCGATGGTAGAACGAAGCGGCGTCGAAGCCTGTAAAGCGGCGGTGCACAATCTTCTCGCGC
AACGCGTCAGTGGGCTGATCATTAACTATCCGCTGGATGACCAGGATGCCATTGCTGTGGAAGCTGCCTGCACTAATGTTCCGGCGTTATTTCTTGATGTCTCTGACCAGACACCC
ATCAACAGTATTATTTTCTCCCATGAAGACGGTACGCGACTGGGCGTGGAGCATCTGGTCGCATTGGGTCACCAGCAAATCGCGCTGTTAGCGGGCCCATTAAGTTCTGTCTCGGC
GCGTCTGCGTCTGGCTGGCTGGCATAAATATCTCACTCGCAATCAAATTCAGCCGATAGCGGAACGGGAAGGCGACTGGAGTGCCATGTCCGGTTTTCAACAAACCATGCAAATG
CTGAATGAGGGCATCGTTCCCACTGCGATGCTGGTTGCCAACGATCAGATGGCGCTGGGCGCAATGCGCGCCATTACCGAGTCCGGGCTGCGCGTTGGTGCGGATATCTCGGTAG
TGGGATACGACGATACCGAAGACAGCTCATGTTATATCCCGCCGTTAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACCGCTTGCTGCAACTCTCTCA
GGGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCTGGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATG
CAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGA
LacI_X86 Mutation (amino acid 61 is changed from TCG to CTG)
ATGAAACCAGTAACGTTATACGATGTCGCAGAGTATGCCGGTGTCTCTTATCAGACCGTTTCCCGCGTGGTGAACCAGGCCAGCCACGTTTCTGCGAAAACGCGGGAAAAAGTGGAAGC
GGCGATGGCGGAGCTGAATTACATTCCCAACCGCGTGGCACAACAACTGGCGGGCAAACAGCTGTTGCTGATTGGCGTTGCCACCTCCAGTCTGGCCCTGCACGCGCCGTCGCAA
ATTGTCGCGGCGATTAAATCTCGCGCCGATCAACTGGGTGCCAGCGTGGTGGTGTCGATGGTAGAACGAAGCGGCGTCGAAGCCTGTAAAGCGGCGGTGCACAATCTTCTCGCGC
AACGCGTCAGTGGGCTGATCATTAACTATCCGCTGGATGACCAGGATGCCATTGCTGTGGAAGCTGCCTGCACTAATGTTCCGGCGTTATTTCTTGATGTCTCTGACCAGACACCC
ATCAACAGTATTATTTTCTCCCATGAAGACGGTACGCGACTGGGCGTGGAGCATCTGGTCGCATTGGGTCACCAGCAAATCGCGCTGTTAGCGGGCCCATTAAGTTCTGTCTCGGC
GCGTCTGCGTCTGGCTGGCTGGCATAAATATCTCACTCGCAATCAAATTCAGCCGATAGCGGAACGGGAAGGCGACTGGAGTGCCATGTCCGGTTTTCAACAAACCATGCAAATG
CTGAATGAGGGCATCGTTCCCACTGCGATGCTGGTTGCCAACGATCAGATGGCGCTGGGCGCAATGCGCGCCATTACCGAGTCCGGGCTGCGCGTTGGTGCGGATATCTCGGTAG
TGGGATACGACGATACCGAAGACAGCTCATGTTATATCCCGCCGTTAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACCGCTTGCTGCAACTCTCTCA
GGGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCTGGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCATTAATG
CAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGA
LacI_I12_X86 Mutation
ATGAAATATGTAACGTTATACGATGTCGCAGAGTATGCCGGTGTCTCTTATCAGACCGTTTCCCGCGTGGTGAACCAGGCCAGCCACGTTTCTGCGAAAACGCGGGAAAAAGTGGAAGCGGCGATGGCGGAG
CTGAATTACATTCCCAACCGCGTGGCACAACAACTGGCGGGCAAACAGCTGTTGCTGATTGGCGTTGCCACCTCCAGTCTGGCCCTGCACGCGCCGTCGCAAATTGTCGCGGCGATTAAATCTCGCGCC
GATCAACTGGGTGCCAGCGTGGTGGTGTCGATGGTAGAACGAAGCGGCGTCGAAGCCTGTAAAGCGGCGGTGCACAATCTTCTCGCGCAACGCGTCAGTGGGCTGATCATTAACTATCCGCTGGATGAC
CAGGATGCCATTGCTGTGGAAGCTGCCTGCACTAATGTTCCGGCGTTATTTCTTGATGTCTCTGACCAGACACCCATCAACAGTATTATTTTCTCCCATGAAGACGGTACGCGACTGGGCGTGGAGCATC
TGGTCGCATTGGGTCACCAGCAAATCGCGCTGTTAGCGGGCCCATTAAGTTCTGTCTCGGCGCGTCTGCGTCTGGCTGGCTGGCATAAATATCTCACTCGCAATCAAATTCAGCCGATAGCGGAACGGG
AAGGCGACTGGAGTGCCATGTCCGGTTTTCAACAAACCATGCAAATGCTGAATGAGGGCATCGTTCCCACTGCGATGCTGGTTGCCAACGATCAGATGGCGCTGGGCGCAATGCGCGCCATTACCGAGT
CCGGGCTGCGCGTTGGTGCGGATATCTCGGTAGTGGGATACGACGATACCGAAGACAGCTCATGTTATATCCCGCCGTTAACCACCATCAAACAGGATTTTCGCCTGCTGGGGCAAACCAGCGTGGACC
GCTTGCTGCAACTCTCTCAGGGCCAGGCGGTGAAGGGCAATCAGCTGTTGCCCGTCTCACTGGTGAAAAGAAAAACCACCCTGGCGCCCAATACGCAAACCGCCTCTCCCCGCGCGTTGGCCGATTCAT
TAATGCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGA
LacI Promoter
CGTTGACACCATCGAATGGCGCAAAACCTTTCGCGGTATGGCATGATAGCGCCCGG
LacIQ Promoter
CGTTGACACCATCGAATGGTGCAAAACCTTTCGCGGTATGGCATGATAGCGCCCGG
LacIQ1 Promoter
AGCGGCATGCATTTACGTTGACACCACCTTTCGCGGTATGGCATGATAGCGCCCGG
-These promoter sequences are taken from Glascock and Weickert 1998
| Part | Forward | Reverse |
|---|---|---|
| lacIQ1 Promoter | |

|
| lacIQ Promoter | 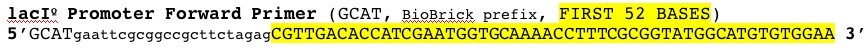 |

|
| LacI_I12 | 
| |
| LacI | |

|
| LacI_X86 (Primers for step 1 of PCR) | |

|
| LacI_I12X86 (Primers for step 1 of PCR) | |

|
Testing Lac promoters and proteins
Construct One
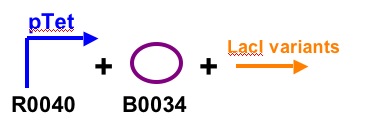
Construct Two
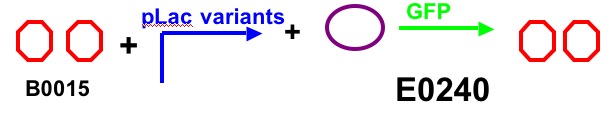
-We will test by ligating each of these constructs together with the 3 different lac promoters and the 4 different Lac proteins. We will measure the output of GFP to characterize the strength of each promoter and protein. We will have 12 constructs all together.