Degradation of Xenobiotics by Halomicrobium mukohataei (Megan Reilly)
KEGG pathways that I have found viable genes for in the H. mukohataei genome:
- Benzoate degradation via hydroxylation
- Styrene degradation
- Caprolactam degradation
Possibly others, but either the gaps in other pathways are due to H. mukohataei possessing an alternate gene rather than one that is already known to exist in other genomes, or that a gene has not been identified in any genomes at all and requires wet-lab experiments to determine the sequence of the gene. These unknown genes must exist in several organisms, because the protein that they produce has been identified.
Contents
Benzoate degradation via hydroxylation
Click here to see the KEGG Pathway
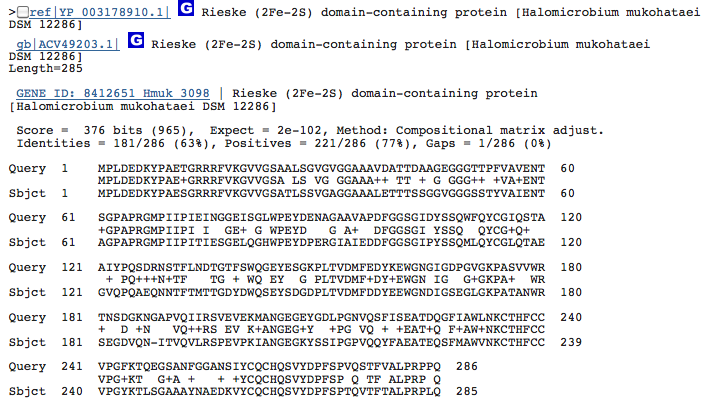
Gene 1.14.12.10 - Rieske (2Fe-2S) domain-containing protein:
(BLAST of H. marismortui; sequence of both microbes obtained from RAST.)
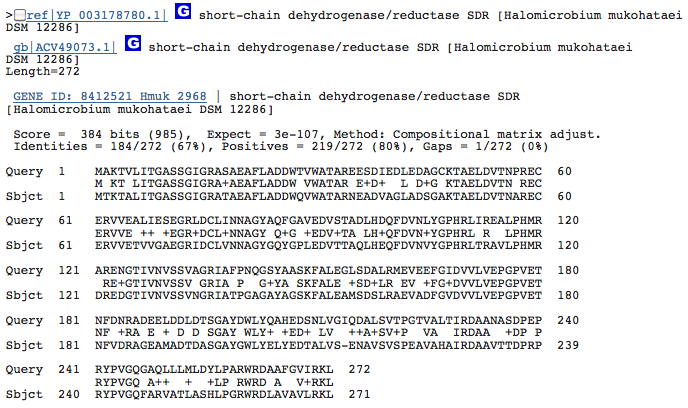
Gene 1.3.1.25 - putatative dehydrogenase
(BLAST of H. marismortui; sequence of both microbes obtained from RAST.)
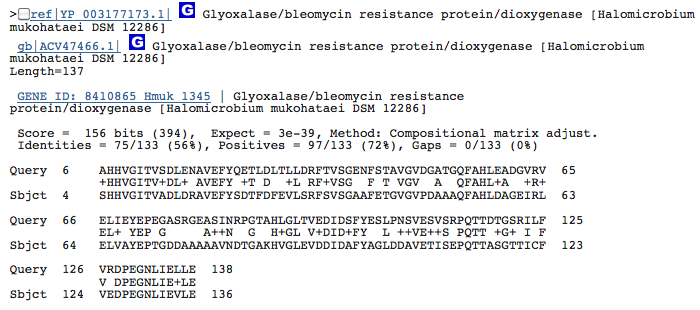
Gene 1.13.11.1 - putatative dioxygenase
(BLAST of H. marismortui; sequence of both microbes obtained from RAST.)
Styrene degradation
Caprolactam degradation
Other potential biodegradable xenobiotics
Benzene, a low molecular weight petroleum hydrocarbon, has been shown to be utilized by an unnamed halophile as its sole source of energy. This organism was also able to degrade toluene, another aromatic hydrocarbon.Fathepure 2006