Difference between revisions of "Designing XOR Gates - two campus approach"
MaCampbell (talk | contribs) (→Davidson XOR Biological Design) |
MaCampbell (talk | contribs) (→Davidson XOR Biological Design) |
||
| Line 12: | Line 12: | ||
''LIST OF Km Values''<br> | ''LIST OF Km Values''<br> | ||
| − | Km of '''3OC12 for LasR''' is 1 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf Egland and Greenberg, 2000]) <br> | + | *Km of '''3OC12 for LasR''' is 1 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf Egland and Greenberg, 2000]) <br> |
| − | Km of '''3OC6 for LuxR''' is 100 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Urbanowski_04.pdf Urbanowski et al., 2004])<br> | + | *Km of '''3OC6 for LuxR''' is 100 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Urbanowski_04.pdf Urbanowski et al., 2004])<br> |
| − | Km of '''LasR* for operator/promoter''' is 11 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/Schuster_04.pdf Schuster et al, 2004])<br> | + | *Km of '''LasR* for operator/promoter''' is 11 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/Schuster_04.pdf Schuster et al, 2004])<br> |
| − | Km of '''LuxR* for operator/promoter''' is 10 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Kapland_Greenberg_87.pdf Kaplan and Greenberg, 1987])<br> | + | *Km of '''LuxR* for operator/promoter''' is 10 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Kapland_Greenberg_87.pdf Kaplan and Greenberg, 1987])<br> |
| − | Km of '''cI dimer for OR1 and OR2''' is 10 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/King_93.pdf King et al., 1993])<br> | + | *Km of '''cI dimer for OR1 and OR2''' is 10 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/King_93.pdf King et al., 1993])<br> |
| − | Km of '''Mnt tetramer for binding to half operator/promoter''' is 50 nM and '''binding whole operator''' is 1 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Berggrun_01.pdf Berggrun and Sauer, 2001])<br> | + | *Km of '''Mnt tetramer for binding to half operator/promoter''' is 50 nM and '''binding whole operator''' is 1 nM ([http://www.bio.davidson.edu/courses/synthetic/papers/Berggrun_01.pdf Berggrun and Sauer, 2001])<br> |
| − | Km of '''Lsr for its binding site''' is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000]) <br> | + | *Km of '''Lsr for its binding site''' is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000]) <br> |
| − | Km of '''AI-2 for LsrR''' is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000]) <br> | + | *Km of '''AI-2 for LsrR''' is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000]) <br> |
| − | Km of '''IPTG for LacI''' is 1.3 µM ([http://www.bio.davidson.edu/courses/synthetic/papers/Gibert_Muller_hill_66.pdf Gilbert and Muller-Hill, 1966]) <br> | + | *Km of '''IPTG for LacI''' is 1.3 µM ([http://www.bio.davidson.edu/courses/synthetic/papers/Gibert_Muller_hill_66.pdf Gilbert and Muller-Hill, 1966]) <br> |
| − | Km of '''LacI for its binding site''' is 10 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/Gilbert_Muller_Hill_67.pdf Gilbert and Muller-Hill, 1967]) <br> | + | *Km of '''LacI for its binding site''' is 10 pM ([http://www.bio.davidson.edu/courses/synthetic/papers/Gilbert_Muller_Hill_67.pdf Gilbert and Muller-Hill, 1967]) <br> |
| − | Km of '''LacI-I12 for its binding site''' is 0.13 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> | + | *Km of '''LacI-I12 for its binding site''' is 0.13 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> |
| − | Km of '''LacI-X86 for its binding site''' is 0.13 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> | + | *Km of '''LacI-X86 for its binding site''' is 0.13 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> |
| − | Km of '''LacI-I12_X86 for its binding site''' is 0.001 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> | + | *Km of '''LacI-I12_X86 for its binding site''' is 0.001 pM (calculated from [http://www.bio.davidson.edu/courses/synthetic/papers/pLac_1980.pdf Schmitz and Galas, 1980])<br> |
| − | '''Half life of molecules in ''E. coli'':''' 2 minutes for mRNA; 1 molecule is 1 nM concentration | + | *'''Half life of molecules in ''E. coli'':''' 2 minutes for mRNA; 1 molecule is 1 nM concentration ([http://www.bio.davidson.edu/courses/synthetic/papers/lsr_thesis_07.pdf Fang Ph.D. Thesis, 2007]); translation rate 15 amino acids per second and transcription is 40 nt per second (from ''Genes VII'' by Lewin). |
== Missouri Western XOR Biological Design 1== | == Missouri Western XOR Biological Design 1== | ||
Revision as of 13:16, 2 July 2008
Contents
- 1 Davidson XOR Biological Design
- 2 Missouri Western XOR Biological Design 1
- 3 Combined Davidson/Missouri Western XOR Biological Design
- 4 Missouri Western XOR Biological Design 2
- 5 Missouri Western XOR Biological Design 3
- 6 Davidson Ampicillin Communication: time delayed cell growth
- 7 Davidson Growth Layouts
Davidson XOR Biological Design
List of auto-inducers and their catalog numbers.
Here is an idea Malcolm and Laurie developed.
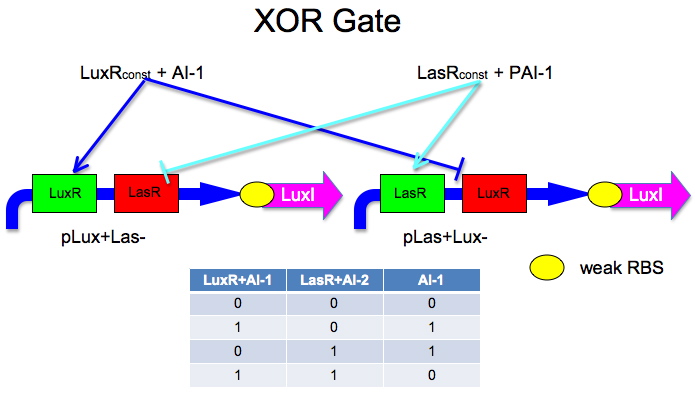
The idea is to have two mirrored halves of the system. LasR is regulated by PAI-1 {3-oxododecanoyl-HSL (3OC12HSL)} and LuxR is activated by AI-1 {3-oxohexanoyl-homoserine lactone (3OC6HSL)}. There is a potential problem in that the Lux half is more likely to get positive feedback than the Las half. This MAY not be a problem because 0/0 is leaky so we put a weak RBS to minimize leaky protein production. Also, if we add AI-2 and AI-1 is produced by leak, then the entire system shuts down. The repressor site is located between -35 and -10 of the promoter. The activator binding site is upstream of -35. This has been documented by Egland and Greenberg
Oligos_to_Build: Sequences we will need to make this XOR gate.
LIST OF Km Values
- Km of 3OC12 for LasR is 1 nM (Egland and Greenberg, 2000)
- Km of 3OC6 for LuxR is 100 nM (Urbanowski et al., 2004)
- Km of LasR* for operator/promoter is 11 pM (Schuster et al, 2004)
- Km of LuxR* for operator/promoter is 10 nM (Kaplan and Greenberg, 1987)
- Km of cI dimer for OR1 and OR2 is 10 pM (King et al., 1993)
- Km of Mnt tetramer for binding to half operator/promoter is 50 nM and binding whole operator is 1 nM (Berggrun and Sauer, 2001)
- Km of Lsr for its binding site is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000])
- Km of AI-2 for LsrR is X nM (www.bio.davidson.edu/courses/synthetic/papers/LuxR.pdf rrrr, 2000])
- Km of IPTG for LacI is 1.3 µM (Gilbert and Muller-Hill, 1966)
- Km of LacI for its binding site is 10 pM (Gilbert and Muller-Hill, 1967)
- Km of LacI-I12 for its binding site is 0.13 pM (calculated from Schmitz and Galas, 1980)
- Km of LacI-X86 for its binding site is 0.13 pM (calculated from Schmitz and Galas, 1980)
- Km of LacI-I12_X86 for its binding site is 0.001 pM (calculated from Schmitz and Galas, 1980)
- Half life of molecules in E. coli: 2 minutes for mRNA; 1 molecule is 1 nM concentration (Fang Ph.D. Thesis, 2007); translation rate 15 amino acids per second and transcription is 40 nt per second (from Genes VII by Lewin).
Missouri Western XOR Biological Design 1
These two XOR circuits are designed to complement each other. Each recieves a cell-to-cell signal (AI-1 or AI-2) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce AI-2. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate.
Design Variables
1. strength on RBS for each of the coding sequences (eg. RBS for enzymes could be weak)
2. order of coding sequences (eg. enzymes could be second for lower expression level)
3. identity of repressors (Davidson is building three different LacI repressors)
4. type of degradation tag on proteins
Things to Do
1. make a list of the parts needed and their building status
2. design, order, and clone 4 hybrid promoters
3. test the 4 hybrid promoters with GFP or RFP outputs
DNA Sequences of Relevant Parts
Oligo design for XOR Hybrid Promoters
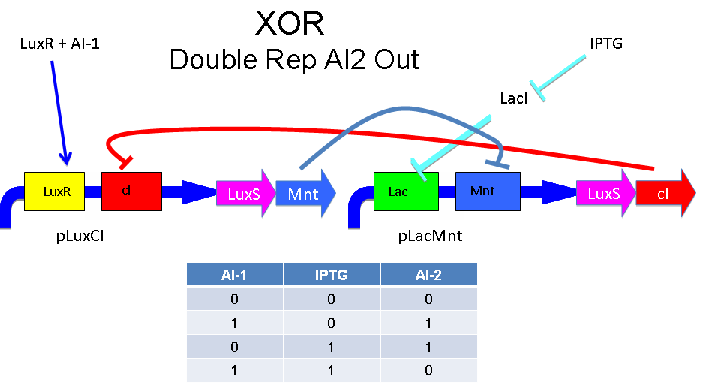
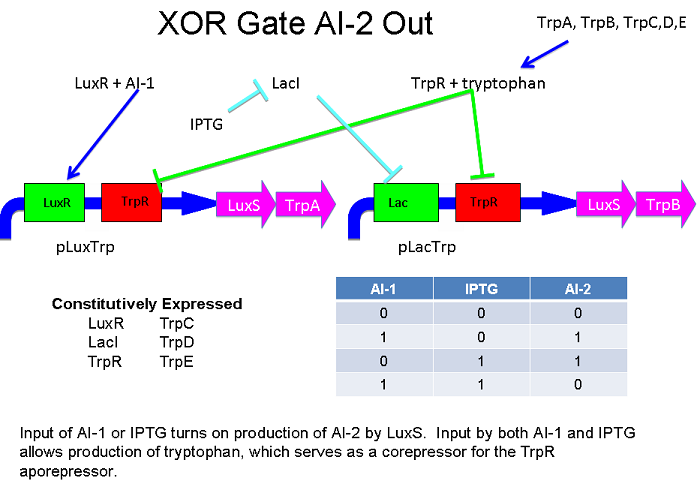
Above - Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed.
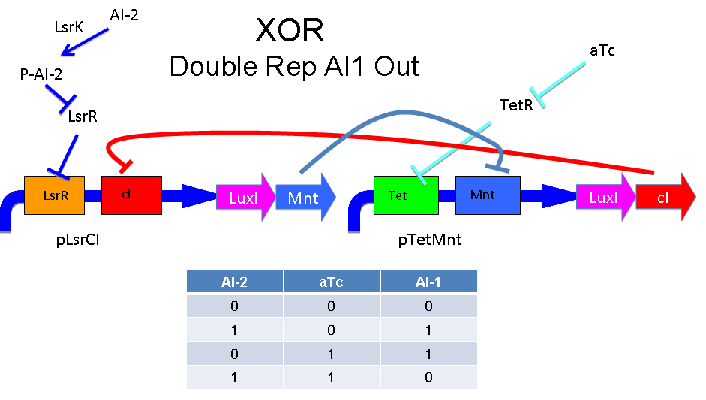
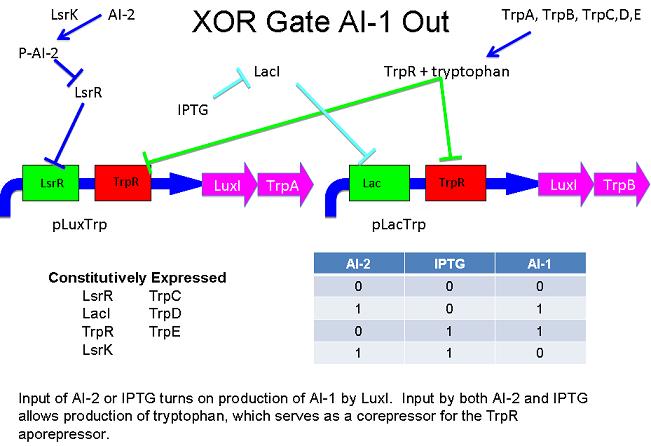
Above - Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.
Combined Davidson/Missouri Western XOR Biological Design
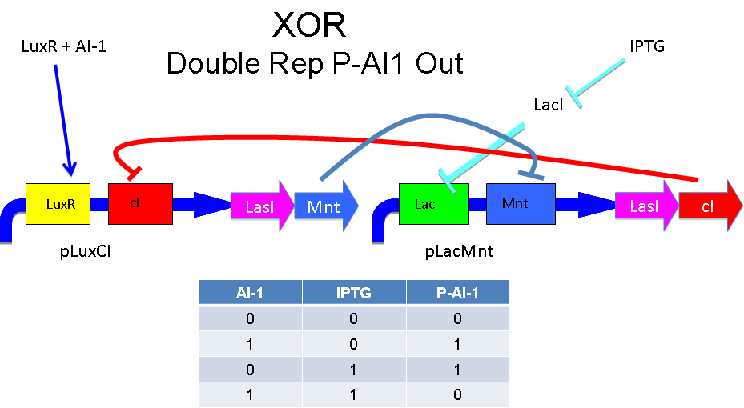
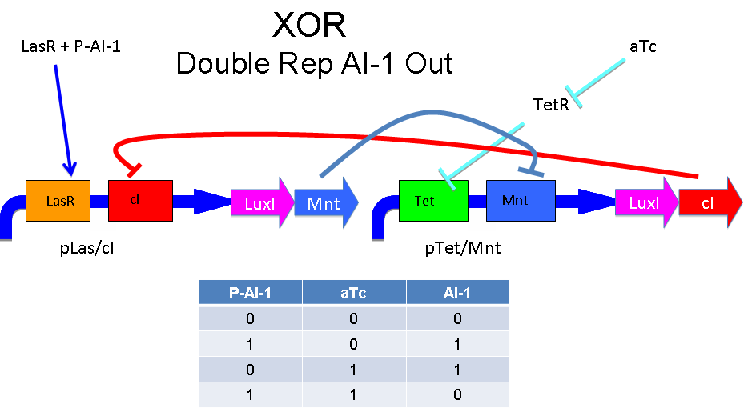
A strength of the Davidson XOR design above is that each of the two signalling molecules used is foreign to E. coli. A strength of the Missouri Western design above is that is uses two complementary XOR gates. The design below combines the two approaches. Each XOR circuit receives a cell-to-cell signal (AI-1 or P-AI-1) and a chemical signal (IPTG or AHL) and processes it into a cell-to-cell signal. Colonies that output AI-1 would alternate with colonies that produce P-AI-1. The input message to be hashed could be encoded by the presence or absence of the chemical signals, which would also alternate.
Input of AI-1 or IPTG turns on production of AI-2 by LuxS. Input by both AI-1 and IPTG allows production of the repressors cI and Mnt, which repress both transcription units. LuxR and LacI are constitutively expressed.
Input of AI-2 or aTc turns on production of AI-1 by LuxI. Input by both AI-2 and aTc allows production of the repressors cI and Mnt, which repress both transcription units. LsrK, LsrR and TetR are constitutively expressed.
Missouri Western XOR Biological Design 2
XOR Based on Tryptophan Anabolism and the TrpR Repressor
Notes: LacI could be the new LacI X86+I12. Also, the output gene should be LuxI, not LuxS. With LuxS, the second cell has to have the Las system in place of the Lux system (JB/AMC).
The idea here is that there are two different XOR gate clones. One takes input of AI1 and IPTG and outputs AI2. The other takes inputs of AI2 and IPTG and outputs AI2. These two clones could be alternated in a pathway of colonies (TE/AG).
Missouri Western XOR Biological Design 3
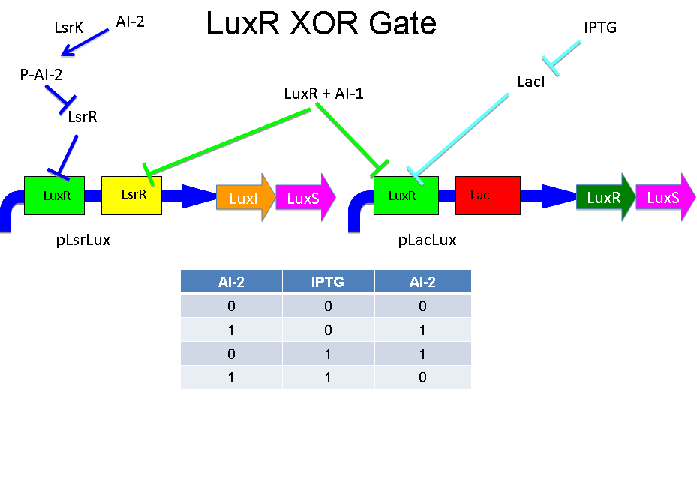
This circuit connects the two transcription units through synthesis of AI-1 and LuxR, which combine to repress both units. It responds to AI-2 and outputs AI-2. An analogous circuit could have input of AI-1 and IPTG and output of AI-1. However, these two could not communicate with each other.
Above - Input of AI-2 or IPTG turns on production of AI-2 by LuxS. Input by both AI-2 and IPTG allows production of LuxI and LuxR, which combine to repress both transcription units. LsrK, LsrR, and LacI are constitutively expressed.
Davidson Ampicillin Communication: time delayed cell growth
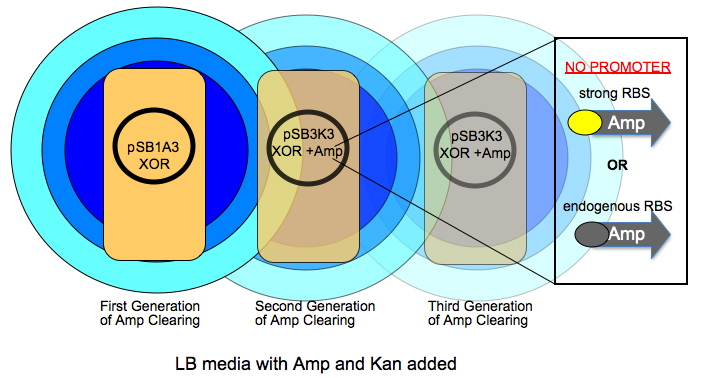
The very first colony has a high copy number plasmid that is Amp and Kan resistant (R). As this colony grows over time, it will digest ampicillin in an increasingly larger circle shown by radiating circles of blue. The subsequent colonies have a lower copy number plasmid and have a promoter-less version of AmpR in addition to the XOR construct. The AmpR coding will have either its native RBS or one we give it.
Davidson Growth Layouts
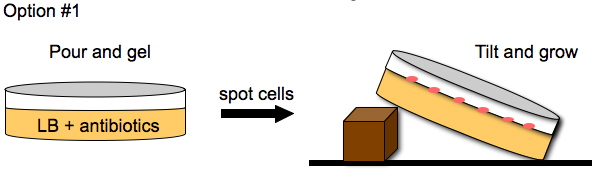
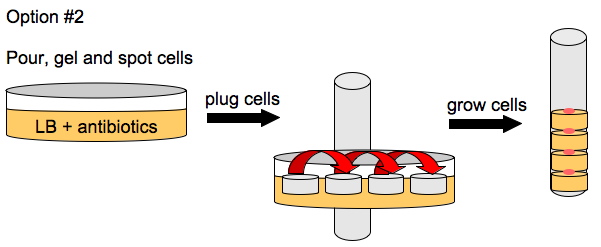
To enhance the unidirectional flow of AmpR, we could either grow the cells on a slant or create a vertical stack of agar plugs. The thickness of the plug would be determined by the thickness of the plates we pour. This may or may not help with the diffusion of AmpR but it is easier to do than microfluidics.