Difference between revisions of "Determining Unique and Conserved Proteins: How to Use Katie's Webpage"
| Line 7: | Line 7: | ||
Next, copy and paste the sequence you want to receive unique and conserved proteins for (still in FASTA format) into the first box. In the second box, add the sequence that you are using to compare your original sequence to. This sequence allows the program to determine which proteins are unique to the first species and which proteins are conserved between the two species. Once your page looks something like this, you're ready to "submit":<br><br> | Next, copy and paste the sequence you want to receive unique and conserved proteins for (still in FASTA format) into the first box. In the second box, add the sequence that you are using to compare your original sequence to. This sequence allows the program to determine which proteins are unique to the first species and which proteins are conserved between the two species. Once your page looks something like this, you're ready to "submit":<br><br> | ||
| − | + | [[Image:Ginandjuice.png]] | |
Revision as of 05:00, 12 November 2009
Firstly, your two sequences must be in FASTA format in order to use our Pairwise Genomic Comparison program. If your sequences are in GenBank format (or another format), visit Claudia's tutorial page first to learn how to convert your sequences to FASTA.
Once you have the two sequences you want to compare in FASTA format, head to the Pairwise Genomic Comparison page. (Note: I'll add the link when Katie finalizes the webpage.)
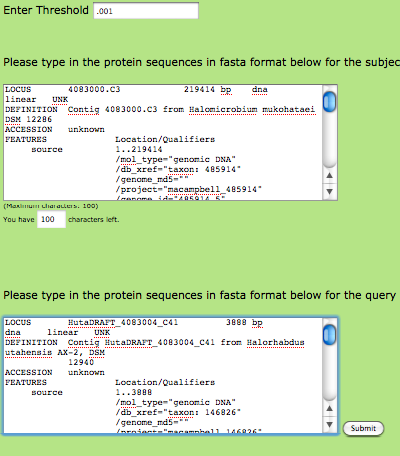
If each of your sequences is less than (Note: Add character limit here) characters, you can use our webpage to perform your comparison. I'll discuss how to download and use the Perl script later on in this tutorial in the event that your sequences are larger than this character limit. Enter your desired Expect (E), or threshold value. Keep in mind that lower E values will be more restrictive and lead to less matches by chance (though any matches found will be more statistically significant). For our purposes, using an E value around 0.001 or 0.01 should be sufficient.
Next, copy and paste the sequence you want to receive unique and conserved proteins for (still in FASTA format) into the first box. In the second box, add the sequence that you are using to compare your original sequence to. This sequence allows the program to determine which proteins are unique to the first species and which proteins are conserved between the two species. Once your page looks something like this, you're ready to "submit":