Difference between revisions of "Dormancy induction"
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | Dormancy is an | + | Dormancy is an essential survival mechanism for most plants. During this phase, plants enter into a state of suspended growth. Environmental factors such as lack of water or extreme temperature change can induce this state until conditions are favorable again. Dormancy pertains to agriculture with the importance of keeping crops alive during harsh weather. |
| + | |||
| + | Plant dormancy is induced, in part, by hormones such as abscisic acid. While it has many uses, abscisic acid inhibits fruit ripening, inhibits cell growth/seed germination, and downregulates enzymes needed for photosynthesis. | ||
[[File:dormancygenes.png]] | [[File:dormancygenes.png]] | ||
| Line 17: | Line 19: | ||
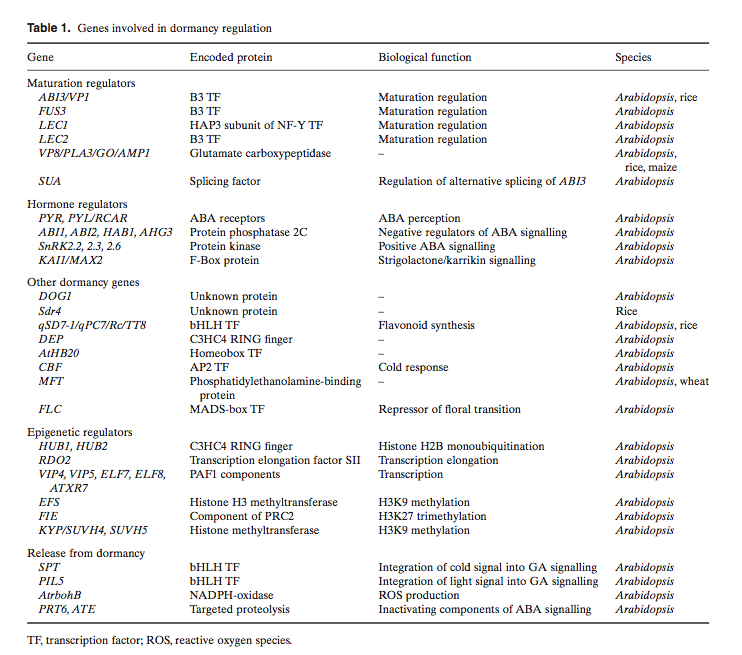
"ABI3, FUS3 and LEC2 encode related plant-specific transcription factors containing the conserved B3 DNA binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001), whereas LEC1 encodes a HAP3 subunit of the CCAAT binding transcription factor (CBF, also known as NF-Y (Lotan et al., 1998)). All four abi3, lec1, lec2 and fus3 mutants are severely affected in seed maturation and share some common phenotypes, such as decreased dormancy at maturation (Raz et al., 2001) and reduced expression of seed storage proteins" - [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3243337/] | "ABI3, FUS3 and LEC2 encode related plant-specific transcription factors containing the conserved B3 DNA binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001), whereas LEC1 encodes a HAP3 subunit of the CCAAT binding transcription factor (CBF, also known as NF-Y (Lotan et al., 1998)). All four abi3, lec1, lec2 and fus3 mutants are severely affected in seed maturation and share some common phenotypes, such as decreased dormancy at maturation (Raz et al., 2001) and reduced expression of seed storage proteins" - [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3243337/] | ||
| − | |||
| − | |||
| − | |||
SUA - splicing factor, splices ABI3- [http://www.ncbi.nlm.nih.gov/pubmed/?term=SUA%20splicing%20factor] | SUA - splicing factor, splices ABI3- [http://www.ncbi.nlm.nih.gov/pubmed/?term=SUA%20splicing%20factor] | ||
| Line 25: | Line 24: | ||
"It is clear from the results that glucose delays germination by activating the ABA signalling pathway via ABI3 and repressing the GA signalling pathway via SPY and RGL2. It has been shown that glucose application has a pronounced effect on ABI3 and RGL2, up-regulating both at the transcriptional level." | "It is clear from the results that glucose delays germination by activating the ABA signalling pathway via ABI3 and repressing the GA signalling pathway via SPY and RGL2. It has been shown that glucose application has a pronounced effect on ABI3 and RGL2, up-regulating both at the transcriptional level." | ||
| − | |||
GAI Della protein [Arabidopsis thaliana] Member of the DELLA proteins that restrain the cell proliferation and expansion that drives plant growth. KEGG - [http://www.genome.jp/dbget-bin/www_bget?ath:AT1G14920] | GAI Della protein [Arabidopsis thaliana] Member of the DELLA proteins that restrain the cell proliferation and expansion that drives plant growth. KEGG - [http://www.genome.jp/dbget-bin/www_bget?ath:AT1G14920] | ||
| Line 45: | Line 43: | ||
ERF4- ethylene-responsive transcription factor 4 - [http://www.genome.jp/dbget-bin/www_bget?ath:AT3G15210] | ERF4- ethylene-responsive transcription factor 4 - [http://www.genome.jp/dbget-bin/www_bget?ath:AT3G15210] | ||
| − | + | PYR/PYL/RCAR - [http://www.ncbi.nlm.nih.gov/pubmed/23265948] - ABA receptor | |
| − | + | DAM genes, Evergrowing ''Prunus persica'': [http://www.ncbi.nlm.nih.gov/pubmed/21899556] [http://jhered.oxfordjournals.org/content/95/5/436.long][http://www.ncbi.nlm.nih.gov/pubmed/21378115] | |
| − | + | SSRs: [[File:All_primers.docx]] | |
| − | [[File: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 18:32, 18 April 2013
Dormancy is an essential survival mechanism for most plants. During this phase, plants enter into a state of suspended growth. Environmental factors such as lack of water or extreme temperature change can induce this state until conditions are favorable again. Dormancy pertains to agriculture with the importance of keeping crops alive during harsh weather.
Plant dormancy is induced, in part, by hormones such as abscisic acid. While it has many uses, abscisic acid inhibits fruit ripening, inhibits cell growth/seed germination, and downregulates enzymes needed for photosynthesis.
Conserved dormancy mechanism- RY repeats in DOG1, Sdr4 promoters
Abscisic acid-insensitive 5-like protein 3 (ABI3). KEGG: [1]
ABI3 papers: [2] [3] Possible link between DOG1 and ABI3- Arabidopsis and B. rapa DOG1 promoter regions contain a RY(arginine, tyrosine) repeat (Graeber et al. 2010) required for ABI3/VP1-mediated expression
LEC1 and LEC2 - [6]
"ABI3, FUS3 and LEC2 encode related plant-specific transcription factors containing the conserved B3 DNA binding domain (Giraudat et al., 1992; Luerssen et al., 1998; Stone et al., 2001), whereas LEC1 encodes a HAP3 subunit of the CCAAT binding transcription factor (CBF, also known as NF-Y (Lotan et al., 1998)). All four abi3, lec1, lec2 and fus3 mutants are severely affected in seed maturation and share some common phenotypes, such as decreased dormancy at maturation (Raz et al., 2001) and reduced expression of seed storage proteins" - [7]
SUA - splicing factor, splices ABI3- [8]
"It is clear from the results that glucose delays germination by activating the ABA signalling pathway via ABI3 and repressing the GA signalling pathway via SPY and RGL2. It has been shown that glucose application has a pronounced effect on ABI3 and RGL2, up-regulating both at the transcriptional level."
GAI Della protein [Arabidopsis thaliana] Member of the DELLA proteins that restrain the cell proliferation and expansion that drives plant growth. KEGG - [9]
RGA1 DELLA protein [Arabidopsis thaliana]: KEGG- [10]
GD1 - [11]
Plant Hormone Signal Induction in Arabidopsis thaliana: [12]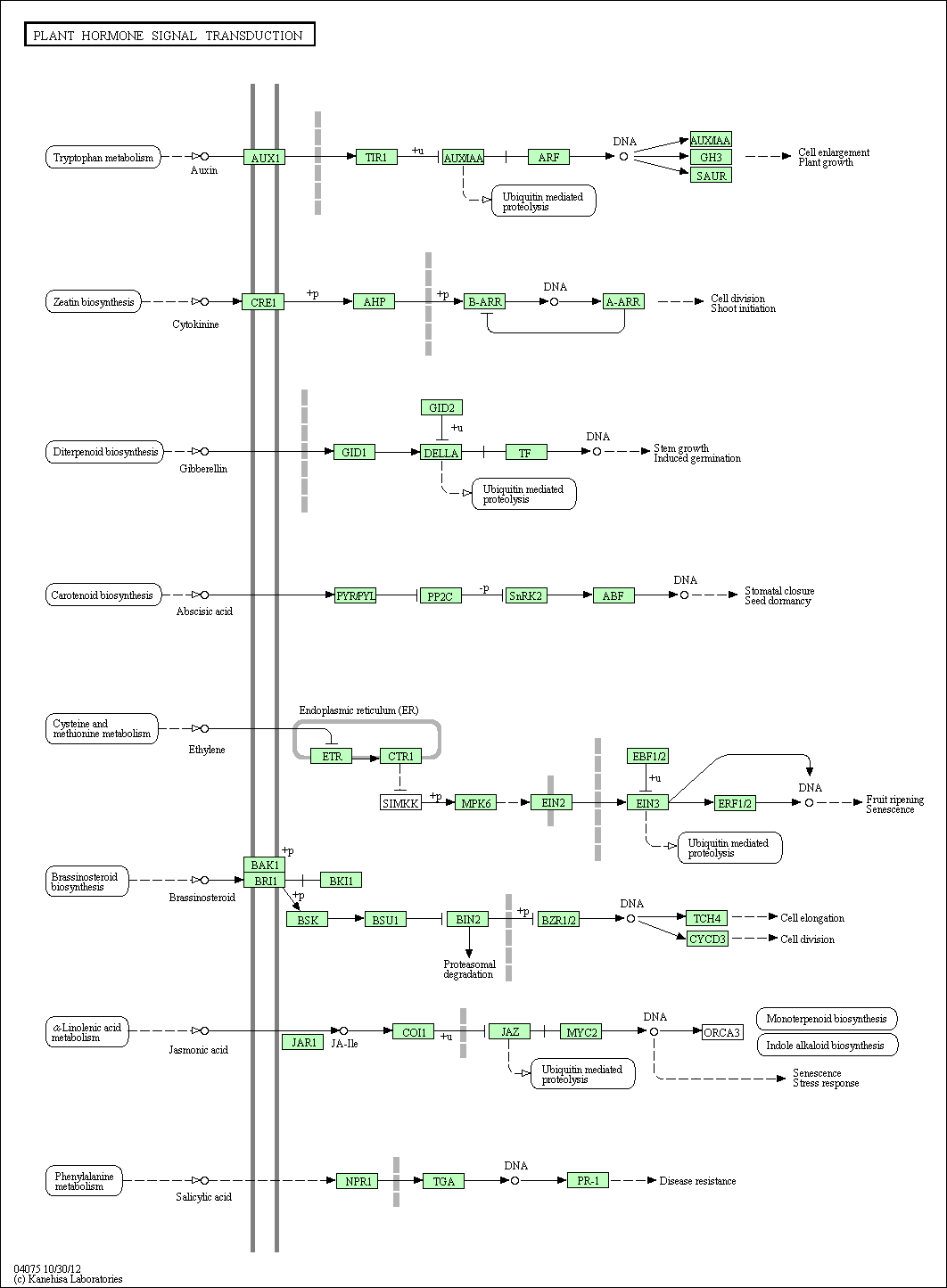
Ethylene- breaks dormancy. etr1 and ein 2 mutants displayed increased dormancy which relates to increased sensitivity to ABA. ctr1 mutations led to decreased ABA sensitivity. [13]
Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar . From this paper: "Three regulatory genes, AP2/EREBP, ERF4, and WRKY11, and one expressed protein gene were associated with this crucial step in dormancy induction (Fig. 7). Based on the function of the respective Arabidopsis homologues, the three regulatory genes could act downstream of ethylene and/or abscisic acid (ABA) signals during dormancy induction." [14]
WRKY11- putative WRKY transcription factor 11- [15]
AP2/EREBP transcription factor - [16]
ERF4- ethylene-responsive transcription factor 4 - [17]
PYR/PYL/RCAR - [18] - ABA receptor
DAM genes, Evergrowing Prunus persica: [19] [20][21]
SSRs: File:All primers.docx