Difference between revisions of "How to deal with multi-named genes"
(→BLAST) |
|||
| (37 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | When investigating a gene, one always must be leery of the possibility that the gene of interest may be referred to by multiple names within the research world. Important Information can be overlooked by assuming a gene only has one name within and across species. This tutorial demonstrates ways to determine if a gene has been assigned multiple names. | |
| + | =='''Finding Your Gene of Interest'''== | ||
| − | ''' | + | Say you just are sitting in Biology class and your teacher mentions ATP synthase has a subunit within its complex that is resistant to oligomyocin called Oligomyocin Sensitivity Conferring Protein. She mentions the gene that encodes the OSCP protein also has the gene name ''OSCP''. This sparks your interest, and you want to investigate this gene more. After class you immediately run home, and go to [http://www.ncbi.nlm.nih.gov/gene/ NCBI Gene] and search for "OSCP." |
| + | [[File:GENE.jpg]] | ||
| − | + | =='''Navigating NCBI'''== | |
| + | [[File:OSCP.jpg]] | ||
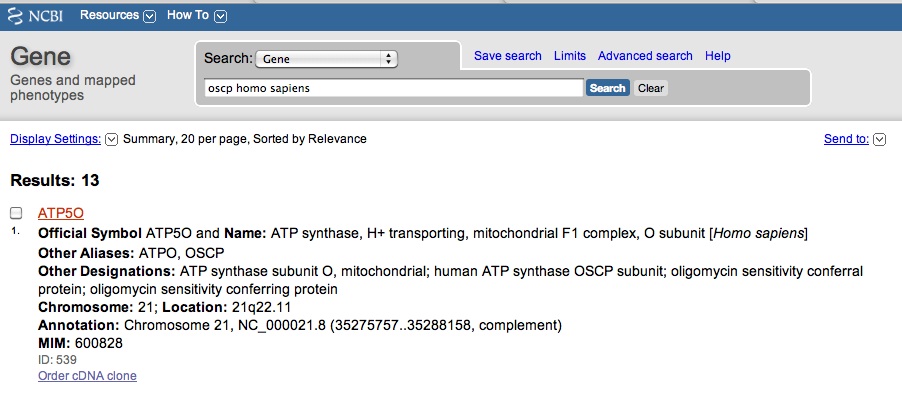
| − | + | Perfect! You get a hit for ''OSCP'' in ''Drosophila melanogaster''. You review the page and with great excitement, you realize you want to research ''OSCP'' in other organisms, including humans. You then NCBI Gene search "OSCP homo sapiens." You get hits, but none specifically say OSCP. The first hit is for a gene called ''ATP50''. | |
| − | |||
| − | |||
| − | Perfect! You get a hit for ''OSCP'' in ''Drosophila melanogaster''. You review the page and realize you want to research ''OSCP'' in other organisms, including humans. You | ||
[[File:ATP50.jpg]] | [[File:ATP50.jpg]] | ||
| Line 19: | Line 19: | ||
| − | '''Is It the Same Gene?''' | + | ==='''Is It the Same Gene?'''=== |
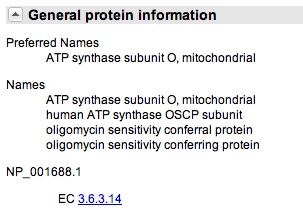
| − | You scroll down to "General Protein Information," as see that gene does encode for oligomyocin sensitivity conferring protein. | + | You click onto the ATP50 link and you scroll down to "General Protein Information," as see that gene does encode for oligomyocin sensitivity conferring protein. This suggest that ATP50 is the ''OSCP'' in humans, just referred to by another name. |
[[File:GPI.jpg]] | [[File:GPI.jpg]] | ||
| Line 28: | Line 28: | ||
[[File:ATP502.jpg]] | [[File:ATP502.jpg]] | ||
| + | ==More Databases for More Verification== | ||
| + | ==='''GeneCards'''=== | ||
| + | The presence of ''OSCP'' in the "Also Known As" section is a little strange because the ''Drosophila'' ''OSCP'' page did not include ''ATP50'' or ''ATP0'' in the "Also Known As" section. | ||
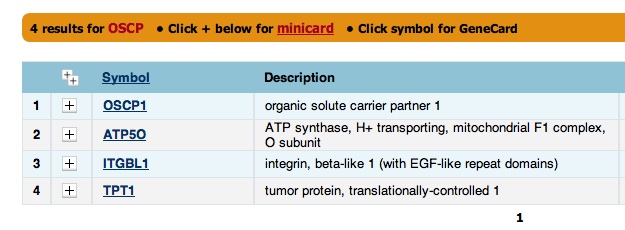
| + | You decide to get further confirmation that ''OSCP'' is ''ATP50''. You search "OSCP" on [http://www.genecards.org/ GeneCards], a database for human genes, and get "ATP50" as a hit. That seems promising. | ||
| + | |||
| + | [[File:GeneCards.jpg]] | ||
| + | ==='''BLAST'''=== | ||
| − | + | You decide you want to find ''OSCP'' in other organisms, but do not want to go through the hassel of searching all of NCBI for alternative names, like you did for Homo sapiens. From NCBI Gene, you again go to the human ATP5O page and find the FASTA nucleotide sequence located under "Genomic regions, transcripts, and products". You can either copy and paste the entire sequence into the BLAST box or use the Ref number. The Ref number in this case is NC_000021.8 for ATP50. This time, you choose to copy and paste the nucleotide sequence. | |
| − | You decide to | + | [[File:FASTA.jpg]] |
| − | |||
| − | |||
| + | Then go to [http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome/ BLAST] and paste the FASTA sequence into the BLAST box. Click BLAST and you compare the nucleotide sequence of ''ATP5O'' against all organisms in the BLAST database. The name of the protein the nucleotide sequence hit encodes is clearly shown, making it easy to detect whether or not you have discovered the ''ATP5O/OSCP'' gene in a species that encodes OSCP protein. | ||
| − | ''' | + | =='''Summary'''== |
| − | + | Genes may have one or more names across species, or even within the same species. As in the case with ''OSCP'' in the ''Drosophila'' page, all of the alternative names for the gene are not always listed, therefore additional investigation can be quite fruitful when it comes to research. Utilize as many database as you can, such a NCBI, Pubmed, GeneCards and BLAST to discover if there is variance in the name of your gene of interest. Knowing all the names a gene has been assigned, will help maximize the amount of information you find about the gene of interest. | |
Latest revision as of 15:59, 24 February 2011
When investigating a gene, one always must be leery of the possibility that the gene of interest may be referred to by multiple names within the research world. Important Information can be overlooked by assuming a gene only has one name within and across species. This tutorial demonstrates ways to determine if a gene has been assigned multiple names.
Contents
Finding Your Gene of Interest
Say you just are sitting in Biology class and your teacher mentions ATP synthase has a subunit within its complex that is resistant to oligomyocin called Oligomyocin Sensitivity Conferring Protein. She mentions the gene that encodes the OSCP protein also has the gene name OSCP. This sparks your interest, and you want to investigate this gene more. After class you immediately run home, and go to NCBI Gene and search for "OSCP."
Perfect! You get a hit for OSCP in Drosophila melanogaster. You review the page and with great excitement, you realize you want to research OSCP in other organisms, including humans. You then NCBI Gene search "OSCP homo sapiens." You get hits, but none specifically say OSCP. The first hit is for a gene called ATP50.
Is It the Same Gene?
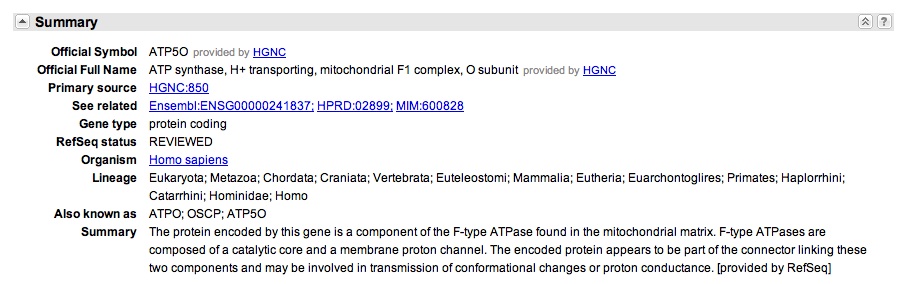
You click onto the ATP50 link and you scroll down to "General Protein Information," as see that gene does encode for oligomyocin sensitivity conferring protein. This suggest that ATP50 is the OSCP in humans, just referred to by another name.
You also notice that there "Also Known As" section, that indicates the gene goes by the names: ATPO; OSCP; ATP5O.
More Databases for More Verification
GeneCards
The presence of OSCP in the "Also Known As" section is a little strange because the Drosophila OSCP page did not include ATP50 or ATP0 in the "Also Known As" section. You decide to get further confirmation that OSCP is ATP50. You search "OSCP" on GeneCards, a database for human genes, and get "ATP50" as a hit. That seems promising.
BLAST
You decide you want to find OSCP in other organisms, but do not want to go through the hassel of searching all of NCBI for alternative names, like you did for Homo sapiens. From NCBI Gene, you again go to the human ATP5O page and find the FASTA nucleotide sequence located under "Genomic regions, transcripts, and products". You can either copy and paste the entire sequence into the BLAST box or use the Ref number. The Ref number in this case is NC_000021.8 for ATP50. This time, you choose to copy and paste the nucleotide sequence.

Then go to BLAST and paste the FASTA sequence into the BLAST box. Click BLAST and you compare the nucleotide sequence of ATP5O against all organisms in the BLAST database. The name of the protein the nucleotide sequence hit encodes is clearly shown, making it easy to detect whether or not you have discovered the ATP5O/OSCP gene in a species that encodes OSCP protein.
Summary
Genes may have one or more names across species, or even within the same species. As in the case with OSCP in the Drosophila page, all of the alternative names for the gene are not always listed, therefore additional investigation can be quite fruitful when it comes to research. Utilize as many database as you can, such a NCBI, Pubmed, GeneCards and BLAST to discover if there is variance in the name of your gene of interest. Knowing all the names a gene has been assigned, will help maximize the amount of information you find about the gene of interest.