IGEM 2009 notebook
Contents
- 1 Roclemente 16:54, 3 June 2009 (EDT)
- 2 Roclemente 17:21, 4 June 2009 (EDT)
- 3 Roclemente 17:13, 5 June 2009 (EDT)
- 4 Roclemente 10:40, 8 June 2009 (EDT)
- 5 Roclemente 11:54, 9 June 2009 (EDT)
- 6 Roclemente 08:46, 10 June 2009 (EDT)
- 7 Roclemente 15:14, 11 June 2009 (EDT)
- 8 Roclemente 22:46, 12 June 2009 (EDT)
- 9 Roclemente 17:24, 15 June 2009 (EDT)
- 10 Roclemente 12:30, 16 June 2009 (EDT)
- 11 Roclemente 14:51, 17 June 2009 (EDT)
- 12 Roclemente 14:00, 18 June 2009 (EDT)
Roclemente 16:54, 3 June 2009 (EDT)
Shamita and I are trying to find suitable reporter proteins to use. Yesterday, Leland and Alyndria were working on ways to insert the gene sequences into the plasmid. Upon seeing how they wanted to manipulated the reporter gene to include the logical clauses, we came up with a few criteria for the reporter genes we would use. The following criteria for genes are listed in order of the broadest aspect to look at to the narrowest aspect:
a) Doesn't contain restriction sites for the 4 restriction enzymes (EcoR1, Xbal, Spel, Pst1) used to cleave the Biobrick part out of the plasmid.
b) Contains 6 cutter restriction sites.
c) These restriction enzymes aren't blunt (cleave straight down at one spot).
d) These restriction sites are close to thge 5' (beginning) end of the sequence.
e) These enzymes are easiest to work with and cheapest.
We are finding the part numbers of the reporter genes we want to use (antibiotic resistance, fluorescence, LacZ) through our own GCAT because we know these ones work. We are then locating these parts on the parts registry [1] website. We copied and pasted the gene sequences we obtained from the registry onto the ApE software [2]. From here, we were able to generate a genetic map of each gene that outlined each restriction site that fit our criteria. We put each genetic map, alongside the part number used, into a Word document:
http://gcat.davidson.edu/GcatWiki/images/0/0e/Restriction_Site_Mapping_on_Reporter_Genes.doc
So it looks like we've changed directions. The team is looking towards the wet lab portion of our research more now. Instead of simply speculating about which reporter proteins we think will work best and which tRNA suppressors to use, we're going to physically test it out ourselves. A few tasks at hand in the second part of this afternoon:
1. What are the DNA gene sequences of the 12 different tRNA suppressors we want to use?
2. What is the sequence of the DNA that is cleaved off of both ends of the tRNA when it is transcribed?
We e-mailed Anderson to find out the exact tRNA gene sequences because we had a hard time finding this information everywhere else. Most papers we found describing the 5-base codon tRNA suppressors simply referred to Anderson's paper [3]. Anderson replied near the end of the day with the gene sequence. According to his email, the only difference between different tRNA genes is the anticodon loop. Therefore, if we just substitute all the anticodon loops in the invariable part of the sequence, we have our different tRNA sequences. We plan on using the Lancilator to help us break these tRNA sequences into smaller oligonucleotides that can anneal at around the same temperature because it is not possible for us to order oligos that are greater than 70 nucleotides long.
Roclemente 17:21, 4 June 2009 (EDT)
I started out the day looking at my time-sensitive group research qustion about reporter proteins. I started making a table listing the advantages and disadvantages of different reporter proteins. Then the group found out that there was a major roadblock to our project: a stop codon was located in our ATG-5mer oligo. The stop codon would have been part of the BioBrick scar. The group looked at several ways to work around this problem. The idea that I looked most into was replacing the restriction sites that were used in the scar through standard assembly. According to the judging criteria for iGEM, I found that a team could possibly alter the standard assembly method as long as they wrote a letter to the iGEM judges explaining our plan in a detailed manner. From there, I set off to understand the BioBrick scar more by making a document highlighting the exact positions of the rsetriction sites on the BioBrick parts and how they were used to put more than one part together. I then found several other pairs of restriction sites that complemented one another and could be used in place of the standard scar between Xba1 and Spe1. At the end of the day, the group got the chance to speak with Dr. Campbell and he suggested a hybrid idea which would combine Davidson's restriction site idea with Missouri's PCR idea. I'm not quite sure of what this idea is exactly yet, but we'll all clarify our vague idea of it tomorrow morning after a talk with Dr. Eckdahl.
Roclemente 17:13, 5 June 2009 (EDT)
This morning we began exploring different options for bypassing the stop codon problem from yesterday. If we wanted to make sure we were using the BioBrick parts in our GCatalog, there was no way we were going to be getting rid of the Xba1 restriction site that contained the "TAG" which codes for a stop codon. Therefore, we proposed looking at restriction sites within the reporter genes instead, therefore addressing the already-placed BioBrick ends. Using the document me and Shamita created on "Restriction Site Mapping on Reporter Genes" (see first journal entry), we determined that the reporter gene with the restriction site closest to the beginning of the sequence was YFP. We proposed cutting the YFP gene that we already had in stock at this restriction site and inserting an oligo (which we would have ordered) with a sequence in the following order: "BioBrick prefix - ATG - 5mer - part of YFP gene to the left of the cleaved restriction site."
After clarifying our own idea with one another, we spoke with some Missouri kids to clarify the said "hybrid" idea which, according to Dr. Campbell, combined the ideas of both campuses. The hybrid idea was based upon our idea of cutting the reporter gene at a restriction site as well, except that our primer would start out with a primer for our reporter gene, and to the left of the primer would be a "tail" consisting of the ATG and 5mer and either an Xba1 or EcoR1 restriction site (We still have to decide this. Our decision will probably be based on the restriction site we end up using in our reporter gene and whether or not that is complementary to either the Xba1 or EcoR1 sites.). We would use the PCR technique and use our in-stock reporter gene strand as our template strand (it will be denatured into single strands) and we will add our oligo. We propose that the primer will synthesize the rest of the YFP strand as well as add on nucleotides to complete the oligo tail. In the end, we will have a double stranded DNA sequence consisting of either the Xba1 or EcoR1 site, the ATG, the 5mer, and the reporter gene. The beauty of this hybrid idea is that we have flexibility as far as the reporter gene we choose to use since all we really need for it is the beginning sequence (for the primer) and a known, "hearty" restriction site. Once we cut this double strand at this restriction site on the reporter gene as well as at the Xba1 or EcoR1 sites, we can insert this into a plasmid that contains the rest of that reporter gene after the restriction site we choose to use and the complementary Xba1 or EcoR1 site.
I made a Word document outlining the promoter and RBS parts we have in our registry on the Davidson campus. Leland and I went through the registry parts on the GCATalog to look for potential promoters and RBS parts. We went through the document as a group and analyzed each of these potential parts on partsregistry.org to see which ones would most likely work the best in the miniprep we'll start preparing for on Sunday evening. Here is the document we wound up with:
http://gcat.davidson.edu/GcatWiki/images/d/d2/Promoters_and_RBS.doc
Roclemente 10:40, 8 June 2009 (EDT)
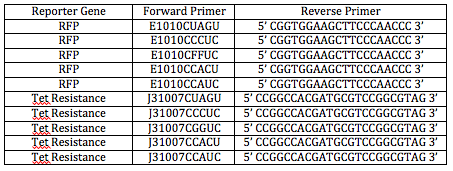
We're starting the day by finalizing the oligos we will be ordering to have synthesized. Two groups of oligos must be sequenced: primers and tRNA gene sequence. The primers will consist of a forward and reverse primer. The forward primer will consist of the following in this 5' - 3' order: GCAT - BioBrick Prefix - ATF - 5mer - first 20 nucleotides in the reporter gene sequence. These 20 nucleotides won't include the ATG that naturally comes at the beginning of the reporter gene because we want to make sure the ribosome attaches to the first ATG and reads the logical clause instead of going straight to the ATG at the start of the reporter gene. The reverse primer will consist of the first 20 nucleotides downstream of the restriction site we plan to use in the reporter gene. I worked on finding the primers for the Tet resistance gene (part J31007) while Lyn worked on finding the primers for the RGP gene. Dr. Campbell decided we should use the BamH1 restriction enzyme because it is very "hardy." The following document outlines the 5 forward primers (each with one of the five variable 5mers) we will order for amplification of the Tet resistance FML:
This is the reverse primer we will order for the Tet resistance FML:
Roclemente 11:54, 9 June 2009 (EDT)
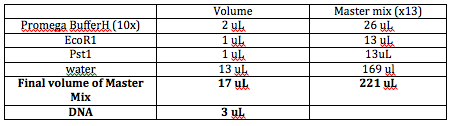
We need to digest the plasmids at the EcoR1 and Pst siets. Everytime we're going to set up a digestion, we'll want the following in these volumes: 10x Buffer (2 uL), DNA (~ 3 uL), enzymes 1 and 2 (1 uL each), water (13 uL), and final volume of all of these put together should be 20 uL.
Some notes about digestion: Enzymes should make up 10% of the final volume. As a general rule of thumb, make sure the tip of your pipet is close to the surface of the solution. This is especially pertinent when gathering 1 microL of each of the enzymes. The most sensitive components are the enzymes, so they should go last.
We'll have a bunch of tubes with each 3 microL of the specific DNA. We'll make a cocktail in a bigger tube consisting of the buffer, 2 enzymes, and water. The rationale behind thissis that you'll be less likely to make a bigger mistake with a bigger volume (versus getting 1 microL of enzyme per specific DNA). You'll also know that there is no problem with the enzyme if all the DNA don't cut. If only one specific DNA doesn't work, then we'll know that it's because there's something wrong with the specific DNA rather than anything in the "Master Mix." Since we have to do 11 digestions, we'll make enough of the master mix for 12 tubes to make sure we have enough. We'll take 17 microL of the Master Mix to each tube of DNA.
The optimal temperature for most enzymes is 37 degreees celsius. You'd like to have enzymes with the same optimal temperatures (for ex. restriction enzymes that start with B usually have an optimal temperature near 50 degrees. You wouldn't want to use both B-enzyme and EcoR1 because EcoR1 would die).
What percent gel should we use?
Look at the Davidson lab protocol on the Davidson-Missouri Wiki. Generally, bigger molecules require an agarose gel with a lower concentration.
What's next in the future?
5' end additions are done by PCR. We'll ampliffy this double-stranded DNA. We'll cut with both EcoR1 and BamH1 or Nco1 (depending on which reporter gene you're using). Then we'll cut the plasmid with the reporter with the same restriction enzymes used.Then, to throw away the remaining DNA that we don't want, we'll run a gel and throw away the smaller band (because the larger band is probably made of the plasmid we want to keep).Then we'll take the double stranded DNA we amplified and the purified plasmid and ligate, transform, and screen them and verify the sequence.
While we wait for the oligos, we can cut the plasmid with reporter on the same restriction enzyme already.
How are we building the tRNAs?
We'll assemble the oligos into genes by mixing our oligos together, boiling them, and very slowly allowing them to cool. This will require many calculations. We'll cut the plasmids with EcoR1 and Pst1. to be ready to receive the enes we're assembling.We'll have to gel purify the plasmids to get them away from their inserts. We'll ligate, transform, and screen the genes and the plasmids and verify the sequence. On paper you shouldn't have two sites cut with different restriction enzymes stick together, but if there's selection pressure, these sites may cut some ends to fit together. This is why we must screen them.
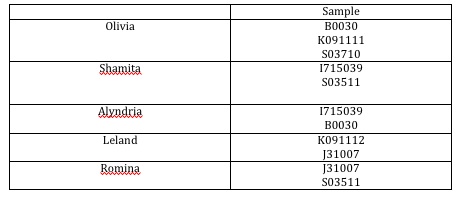
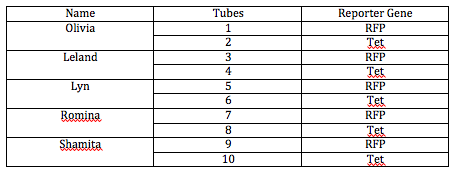
After Dr. Campbell taught us all of the above, the 5 biologists put together a document outlining exactly how we would go about cutting the two sets of plasmids (one set will be cut with Eco R1 and Pst1 and othe other set will be cut with Eco R1 and BamH1 or Nco1). The following table outlines which biologist was going to work with which sample of DNA:
The following summarize the diferent materials and volumes of these materials that we'll need for our digestion of the plasmids:
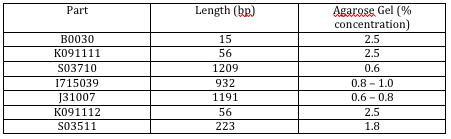
This last table highlights the agarose percent concentration we should use for each part according to how many base pairs it contains:
The biologists digested the plasmids that we plan on using once our oligos arrive in a day or two. Afterwards, we made the two agarose gels that were most compatible with the parts we were going to run. This gel is going to purify the plasmids so that we have the parts that we're going to use later on, minus the rest of the DNA in the plasmid that we don't need. Before physically creating the gel, we figured out how much agarose we would need to make our two different agarose gel concentrations, 0.8% and 2.5%. Next, we made sure the mold and the big teeth were in place. We used the big teeth at the end of the mold because we plan on running 20 uL of DNA versus a running a smaller portion which would require us to use small teeth. We then measured the amount of agarose that we calculated we would need, placed this in a volumetric flask greater than 60 uL to allow for fizz to rise up, then measured out 60 uL of buffer and mixed this in the same flask with the agarose. We placed clear plastic wrap on top of the flask and heated it up for 1 minute and 20 seconds in the microwave. After we heated it, we took it out and checked to see if there was still agarose present that hadn't yet melted. There was, so we placed it back in the microwave and heated it for another 20 seconds. All the agarose had melted. We added 1 uL of EtBr (ethidium bromide) to the solution and quickly poured this into the mold. It needs about 15 to 20 minutes to cool. Dr. Campbell made the agarose gel that was 2.5% concentrated gel while we watched. We then made the 0.8% concentrated gel while he watched.
The following table lists the order in which we loaded our DNA into the gel. The first column all the way to the left is represented by number 1:
Roclemente 08:46, 10 June 2009 (EDT)
After about 30 minutes after we began the gel electrophoresis, we looked at our gels with UV light to see how far the inserts had travelled and where the plasmids were. Then we took each gel to be photographed in the freezer room (231). We used the following DNA length marker to measure how long each sequence represented by each band was:
We compared our data to the lengths we expected to get according to the registry online. The first gel we photographed was the one that was 2.5% concentrated:
The first two columns of the DNA we ran, both consisting of B0300, were fairly the same as the length we expected. However, the last two columns, consisting of K091111 and K091112 respectively were supposed to be 86 bp long, however, according to our data, K091112 was longer. Leland and I looked at Pallavi Penumetcha's paper which delved more closely into these two parts and what she wrote in her paper and what she entered into the registry conflict. We are currently corresponding with her to figure out the exact discrepancy.
We then photographed the 0.8% gel:
The length of the sequences in almost every column matched up with the lengths we expected. Only the 4th and 5th columns didn't come out clearly. We think that the DNA inserted in column 5 weren't inserted properly and diffused on its own. We couldn't see bands in the 4th column at all until we adjusted the color a bit on the photographer and saw that column 4 was pretty much identical to column 5. We assumed that column 5 bled into column 4.
Roclemente 15:14, 11 June 2009 (EDT)
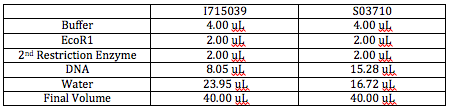
We are digesting two parts today: I715039 (pLac+RBS+RFP) and S03710 (Tet Resistance). We want both the plasmids and the inserts in each case. We are aiming for a final volume of 40 uL for each part. This means that each digestion will consist of 4 uL buffer, 2 uL EcoR1, and 2 uL of their other respective restriction site (BamH1 for S03710 and Nco1 for I715039). The only parts that will vary are the amount of DNA and water we use. The amount of DNA we use depends on the concentration of the DNA we have (found through use of the NanoDrop). Since we want 2500 ng of DNA, we can use the concentration of DNA to figure out how much volume of this concentrated DNA we'll want to use in your digestion. If we subtract the volumes of buffer, two restriction enzymes, and DNA we plan on using from 40 uL, we'll have the amount of water we need to add. Here are the different volumes of materials we plan on using in our double digestion:
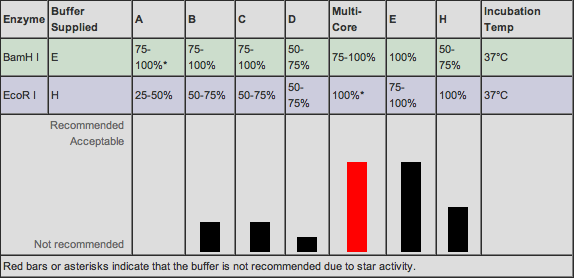
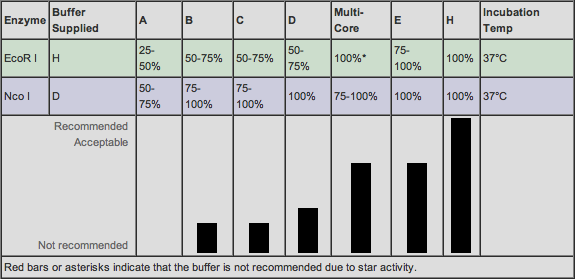
Now that we know how much of each material we were going to use, we needed to find out which buffer would be compatible with each sample according to the restriction enzymes we were using and how effective they'd be in different salt solutions. Online on the Promega "Compatible Buffers" website, we were able to find which Promega buffers would be most compatible when doing a double digest with EcoR1 and BamH1:
Here is a chart noting buffers and their compatiblity with EcoR1 and Nco1:
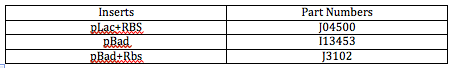
We still have to wait for the restriction enzymes to arrive, so afterwards, we transformed three different inserts containing needed promoters and RBSs into vectors. We used the Zippy Transformation method. These promoters and RBSs will later be added to the FMLs once they are already inserted into the reporter cells. The following table outlines these inserts and part numbers:
The restriction enzymes have arrived. We will now be able to do PCR to amplify the RFP gene (I715039) and the Tet resistance gene (S03710) as templates. We will be using the primers which have also arrived. We ordered forward and reverse primers so that our amplified sequence will consist of the following in this order:
GCAT-ATG-5mer-Reporter Gene
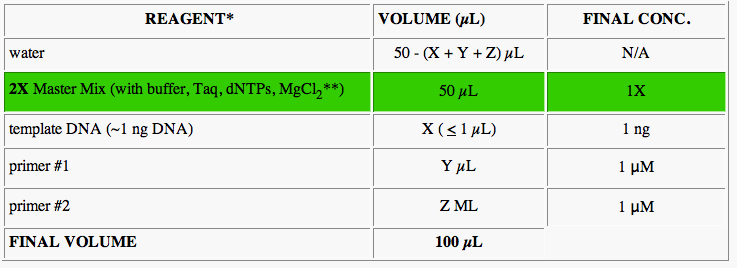
The following table lists the materials will be needed for this PCR technique which uses the "Green Promega master (Monster) Mix:"
This is the PCR procedure:
These are the variables that we will be adding to each separate sequence we want to be amplified:
This table shows how we labeled the tubes based on how we divided the PCR work for the 10 tubes:
Now we are giong to assemble the tRNA oligos together to form the tRNA genes. We could only order a certain length of sequence and the tRNA genes outdid these specified lengths. We used the Lancilator to find various oligonucleotides that overlapped and would be able to anneal together to form the tRNA genes. We used the Davidson lab protocol entitled "Buiding dsDNA with Oligos" to assemble the oligos:
Roclemente 22:46, 12 June 2009 (EDT)
We had two main goals for today: 1. perform a tRNA ligation and transformation 2. perform two electrophoreses: one to verify the PCR sequences we obtained and another to purify the digested reporter plasmids we would need
We began the day by pouring the agarose gels according to which concentration of buffer would be most compatible with the molecular weight of the substance we wanted to isolate. For both the digestion of E01010 and J31007, according to the "Optimize your Gel" website found on the Davidson Lab Protocols site for iGEM 2009, we wanted a 0.4% concentrated gel. For the PCRs, we wanted a 1.6% concentrated gel to optimize our retrieval of the Tet gene and a 1.4% concentrated gel to optimize our retrieval of the RFP gene.
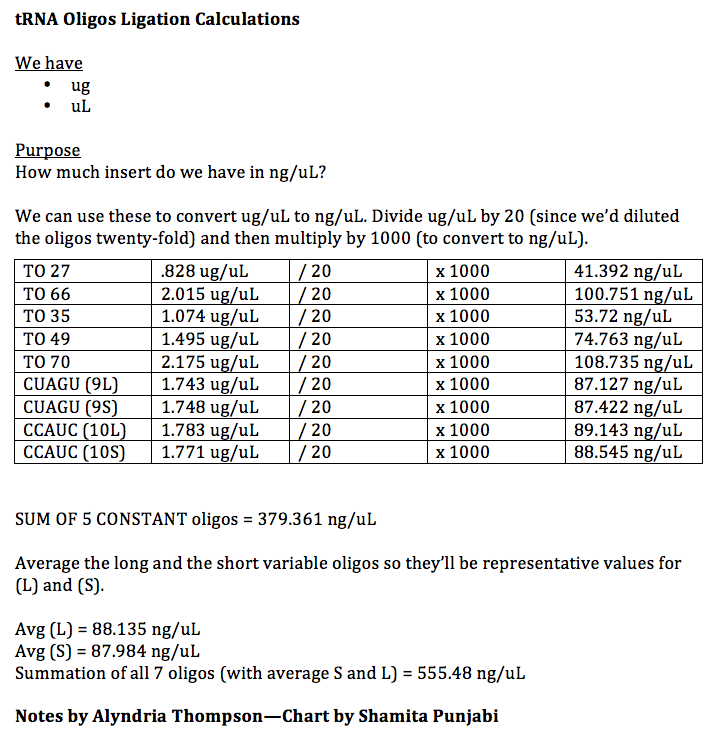
While we waited for the gels to harden for about 30 minutes, we began calculating how much of each material we would need to ligate our tRNAs. We used the following equation to find out how much insert we should use:
Using the lengths of sequences we would be ligating, this is the mass that we needed:
X nanograms of insert = [(2)(144bp)(50 ng linearalized plasmid)]/2038bp X = 7.0657 ng of insert
The following table outlines our calculations for finding the mass of tRNAs we had:
We would attain this mass of tRNA by pipetting an unknown amount of it. In order to attain a more "pipetable" amount of tRNA, we diluted our initial concentration 50X. Since we knew we wanted our final mass to be 7.065 ng, we calculated the volume we would need to take from this new, diluted sample:
DILUTION: 555.48/50 = 11.012 CALCULATION: 11.012 ng/ 1uL = 7.065 ng/ Y uL Y = .64 uL of insert
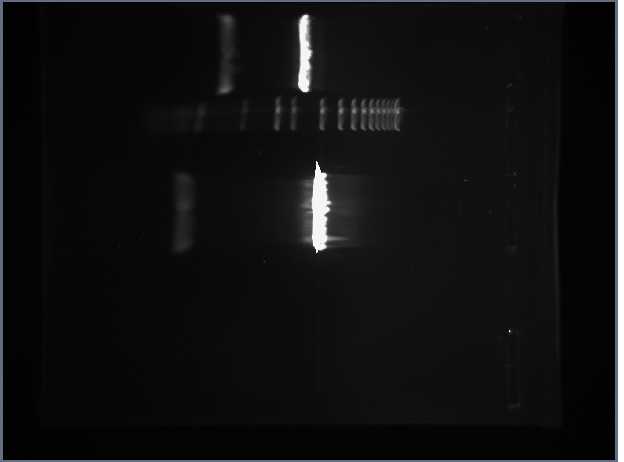
Afterwards, Olivia and Alyndria began the actual ligation process while Shamita, Leland, and I filled the wells of the two gels. The first included the two digested reporter parts. We also included a molecular weight marker. Here is a photograph of this gel:
According to our prior calculations, the Tet plasmid should have been 2958 base pairs long and the RFP plasmid should have been 2380 base pairs long. Our expectations were verified by the data. Shamita and I then sliced the part of thsi gel containing both plasmids and put these slices away to purfiy on Monday.
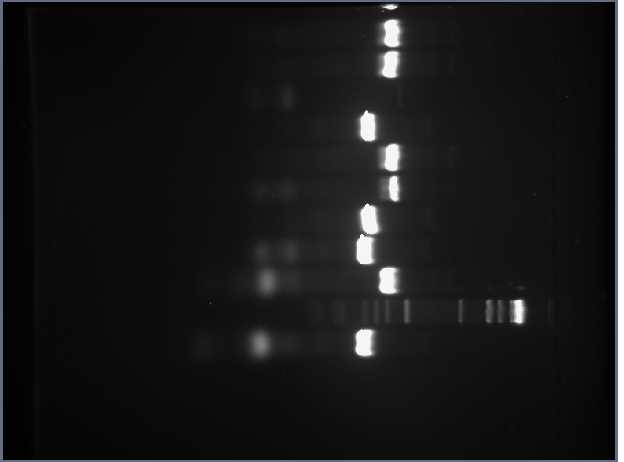
The second gel contained aliquots of the PCR we had done to attain 10 FML sequences: 5 containing the Tet resistant reporter gene and each of the 5 frameshifts that Davidson was assigned and the other 5 containing the RFP reporter gene and each of the 5 frameshifts as well:
According to our data, each sequence containing the Tet resistant gene should have been 317 base pairs long and each sequence containing the RFP gene should have been 446 base pairs long. Our data supports our hypothesis, however, the 3rd well was not successful. This well contained the FML consisting of the Tet resistant gene and the frameshift mutant CUAGU. Before leaving work for the day, Shamita and I prepared and performed PCR using two parts containing the tet resistant gene, J31007 and S03710. We wanted to use both to make sure at least one of them works since we could technically use either of them, even though when we first did PCR we only used the S03710 part. Using the known concentrations of each of these parts, and knowing that we needed 1 ng of each part to include in our PCR samples, we calculated the volume of each 100X-diluted solution we would need:
S03710: 1.636 ng/uL = 1 ng/ X uL X = 0.611 uL
J30017: 0.776 ng/uL = 1 ng/ Y uL Y = 1.289 uL
We will run another gel with both of these parts on Monday and hopefully at least one of them works.
Roclemente 17:24, 15 June 2009 (EDT)
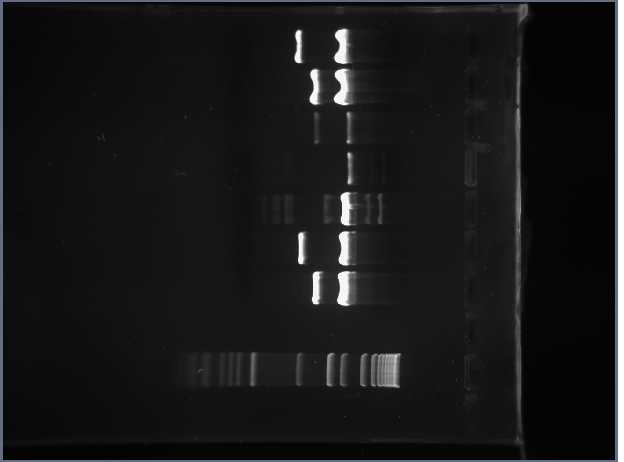
Each biology member of the Davidson iGEM team has been assigned a 5mer to carry on throughout the rest of their respective processes. I've been assigned the 5mer CUAGU. We verified the PCR of the CUAGU FML containing the RFP gene on Friday. However, we weren't able to verify the CUAGU FML containing the tet resistance gene when we ran it on a gel. Shamita and I redid PCR for two parts containing the tet resistance gene: J31007 and S03710, labeled 2* and 2! respectively. I'm running another gel to verify either or both of them. Both sequences should be 317 bp long (this includes the ATF, logical clause, and the tet resistance gene to the left of the BamH1 restriction site). Here is the gel run we collected. The first column represents the 1 kb DNA marker, and the next two are 2* then 2!:
The data shows that the PCRs of both parts wound up at the length we expected. The first time we did PCR on J31007, we must have left a solution out of our procedure on accident. I combined both PCRs together since they are the same length and match the length we expected to get.
Next, we cleaned and concentrated our amplified DNA sequence (#D4014 from Zymo Research). I cleaned the two pieces of DNA that contained the 5mer CUAGU. I labeled the tube with DNA also containing the RFP gene as "1" and the tube with DNA also containing the tet resistance gene as "2!". We needed 2 volumes of buffer for every 1 volume of DNA we added. Therefore, for tube "1," I added 95 uL DNA and 190 uL buffer. I added more DNA to tube "2!" since I combined the DNA from two PCR events to one (2* and 2! from earlier).
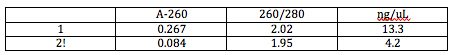
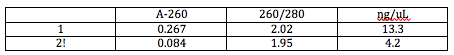
After cleaning the DNA, we found its concentration using the Nanodrop. In a solution made up of 0.5 uL of the DNA and 2.5 uL of TE, we found the following data from the DNA in this solution:
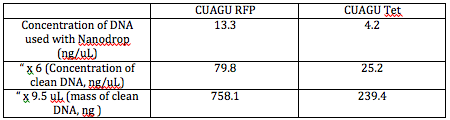
The concentration of the DNA we cleaned was the concentration given by the Nanodrop multiplied by 6 (the DNA was dilute 6-fold for the Nanodrop). We used this concentration and the volume of clean DNA we knew we had (9.5 uL) to find the mass of clean DNA we had:
Before doing digestion, we used the ligation equation (see "Ligation Protocol" in "Davidson Lab Protocols") to figure out how much mass of the DNA insert we would need later on when we did ligation. For the RFP FML, we found we would want the following mass of insert:
ng of insert = (2)(443)(50)/(2320) = 19.0948 ng
We multiplied this mass by 50 because we're counting on losing DNA between the digestion and cleaning we would need to do leading up to the ligation protocol.
19.0948 ng x 50 = 954.741 ng RFP FML
We found the mass of Tet FML insert we would need for ligation as well:
ng of insert = (2)(314)(50)/(2958) = 10.6153 ng
We multiplied this mass by 50 too:
10.6153 x 50 = 530.764 ng Tet FML
Unfortunately, nobody had enough of their clean DNA to satisfy our careful criteria. However, we would need to do this calculation later on so it was helpful we did it anyway. Instead, we just used all the DNA we had, or 9.5 uL for each reporter gene. Since there were 4 people who needed the same master mix, we made 5 parts of the master mix using the following volumes:
After realizing that we would have to wait a day for the digestion to take place, we decided to culture every tRNA cell that colonized. The tRNA that we transformed on Friday did not have a good yield, so last night a few people came in and transformed more tRNA cells. We got a few more colonies. To be safe and make sure we cultured each tRNA, we picked every colony visible to us in both the old and new plates. Using our own given tRNAs, I counted the amount of colonies that grew in the plate we transformed on 6/12 and 6/14. There were 4 colonies that grew on both plates so I labeled a tube for each of these. I awas also in charge of the negative control. There were 2 colonies that grew on the negative control plate so I labeled 2 tubes for that. We will be placing them in an incubator overnight.
Roclemente 12:30, 16 June 2009 (EDT)
We checked on the cells we cultured overnight. Of the cells that I cultured (all containing the CUAGU 5mer and the two negative controls), both negative control tubes worked as well as the tubes I labeled 1 (6/14), 5 (6/12), and 7 (6/12). The rest of mine were either red, meaning the plasmids hadn't ligated properly) or the colonies we used were no good. I took these 5 tubes, along with 3 of Olivia's tubes filled with cells that were cultured (tubes 11, 12, and 13) and miniprepped them to isolate the plasmids from the cells. There was no need to colony PCR to screen for successful ligations because this is only used to know which ones to let grow overnight and we had already done this.
As soon as I finished miniprepping them, I had to leave the lab for a little bit, so Olivia took each my miniprep tubes besides the negative controls and Leland took my negative controls. I completed the protocol for digestion of the tRNAs with EcoR1 and Pst1 for Alyndria's tubes when I got back because she had left a little earlier for lunch.
Afterwards, Shamita and I poured a 1.9% concentrated gel to load the digested tRNAs on. We want to verify that these tRNAs are the appropriate size for if they were cut properly. Since we will have to load 37 wells, and the maximum number we can load on one gel is 18 since 2 wells will have to be used by the molecular weight marker, we needed to pour 2 gels and we will also be using a half of a 2.0% agarose gel that was left over from another experiment.
Shamita and I also made new 0.5X running buffer gel. We did this by adding 450 uL buffer and 9 uL ethidium bromide (1 uL EtBr : 50 uL buffer). Alyndria, Leland, and Shamita loaded these gels.
Olivia and I cleaned our PCR DNA using the "Ethanol precipitate DNA (short protocol)" found in the Davidson Lab Protocols link. To begin with, I added 180 uL dH2O to my two samples, CUAGU with RFP and CUAGU with Tet, to bring their volumes up to 200 uL. During one of the last steps of this protocol, when Olivia and I dried out our DNA using the SpeedVac for a period of 10 minutes, the top of my tube containing the CUAGU 5mer and RFP gene broke. I wasn't sure if a pellet was in there, but I still poured out the ethanol and went on with the protocol. At the end, I resuspended what may or may not have been my DNA in 10 uL distilled water. I NanoDropped it afterward and attained the following data concerning both tubes:
According to this data, any absorption and concentration we may have attained is insignificant because the A-260 must be between 0.05 and 1.50 to be significant and neither of the DNA is. Tomorrow I will do PCR for both FMLs over again.
Roclemente 14:51, 17 June 2009 (EDT)
I found out that my tRNA DNA that I labeled "1" and which contains CUAGU was verified from the gel run Olivia did yesterday (the gel run also included her tRNA DNA samples). At the start of the day I plated this tube of cultured cells again to grow overnight.
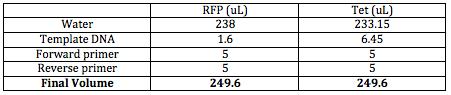
Since PCR will take about 3 1/2 hours, I attempted this procedure at the beginning of the day. I plan on making 5 PCR products for each reporter gene since I now know the environmental conditions are correct. I labeled the RFP tubes 1-5 and the Tet tubes 6-10. I first needed to figure out how much of the template DNA I would need in order to have 1 ng template DNA. I made the following calculations to find this out:
I wanted to add the Monster Mix last, so I made the following table, adding everything else besides the Monster Mix for 5 of the same reporter gene to one tube:
Unfortunately, when I actually mixed the solution together, I forgot to dilute the template DNA. Therefore, we have 100X more template DNA than we would have had we diluted the template DNA. The PCR can still work, but we will have to constantly remind ourselves that there will be extra template DNA in our solution and that this may skew, however slightly, our results later on.
While I waited for the PCR product, I refocused on getting my tRNA DNA ready for sequencing. Using the NanoDrop, I found the concentration of my miniprepped CUAGU DNA that was verified to be 61 ng/uL. The pre-sequencing protocol calls for 60 ng DNA, so I extracted 1 uL from this tube (the extra DNA will be insignificant in the sequencing procedure). I pipetted this into another tube and used the SpeedVac to dry it out. This is how we will send our DNA to be sequenced.
The following is a summary of steps I put together to help me plan out what I will do after the PCR procedure is over (it will be done at 5 pm today):
1. Combine the five matching tubes into one tube. 2. Run each PCR product on a gel to verify that our expected sequence has been properly amplified. 3. Clean and concentrate plasmids so salt solution is optimal for digestion. 4. Double digest PCR products with EcoR1 and BamH1 or Nco1. 5. Clean and concentrate again to get rid of restriction enzymes. 6. Nanodrop inserts in order to do ligation calculations and verify that sequences are present. 7. Ligate FML inserts with purified plasmids. 8. Transform plasmid into cells.
Since I won't have time to run a gel and take a picture once the PCR procedure is complete (we get out at 5:30 and it will be finished at 5:00), I have begun to prepare the agarose gel for use tomorrow. The size of the band I expect to find for the Tet FML is 349 bp (including the GCAT, BioBrick prefix, ATG, 5mer, and reporter gene until about 20 nucleotides or so past the restriction site we will cut at) and 474 bp for the RFP FML. According to the "Optimize Your Gel" link on the Davidson Lab Protocols page, a 1.6% gel will optimize how well I see my Tet bands and a 1.3% gel will optimize how well I see my RFP bands. I can use a gel with a concentration in between both of the optimal concentrations for Tet and RFP so that I can use one gel for both. Therefore, I will use a 1.45% gel. 1.45% of 60 uL buffer solution tells me that I should measure out 0.87 g agar for this gel.
Roclemente 14:00, 18 June 2009 (EDT)
I checked on my PCR products this morning. Tubes 1 and 2 (both containing the RFP FML) had solution scattered on the tube's cover and walls so I placed them in the centrifuge for a bit. Unfortunately, I forgot to put these 500 uL tubes in holders when I centrifuged them so these tubes were destroyed. This was okay though because I was just going to combine each respective FML into one tube. I would have 300 uL of RFP FML PCR product instead of 500 uL and I put all of this in tube 5. I still had 500 uL of Tet FML PCR product left and put all of this in tube 10.
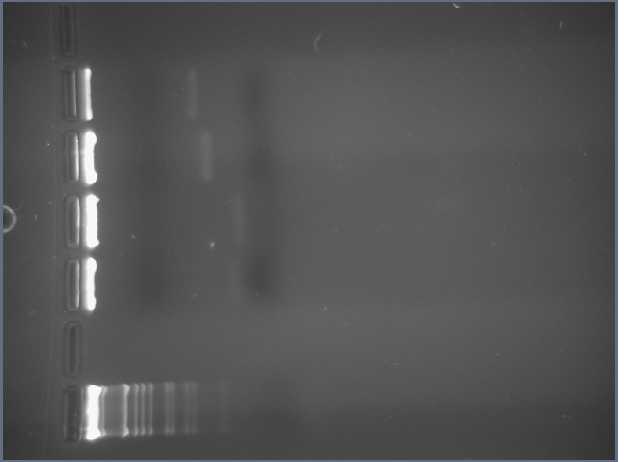
I ran 5 uL of both RFP and Tet FMLs on a gel, as well as a 1 kb molecular weight marker. The MW marker is in lane 1, the RFP is in lane 3, and the Tet is in lane 4 (Leland ran his PCR product in lane 2):
I was expecting the RFP PCR product to be 474 bp long and the Tet PCR product to be 349 bp long. Both appear to come out a bit shorter, but Dr. Campbell and I agreed that we wouldn't count these sequences out because the bands were quite close to what we expected.
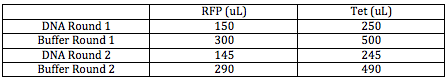
I proceeded to clean and concentrate these two sequences using the procedure outlined in the "Clean and Concentrate DNA (after PCR, before digestion)." Now I had 295 uL of the Tet sequence and 495 uL of the RFP sequence. Since the procedure says I can only have 800 uL in the column at a time, with DNA:Buffer at a ratio of 1:2, I mixed the two in a column in a series of two rounds using the following volumes:
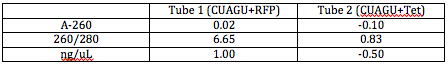
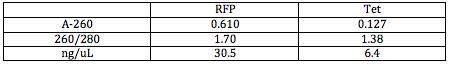
I Nanodropped my DNA after cleaning it and attained the following results:
I wanted the OD260 (Optical Density at 260) to be under 1.5, which it is for both. I lost quite a bit of Tet DNA in the process, and will only continue to lose DNA. To account for this loss of DNA, Dr. Campbell suggested it would be a good idea to heat inactivate the restriction enzymes to get rid of them after digestion instead of cleaning and concentrating the DNA over again.
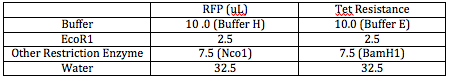
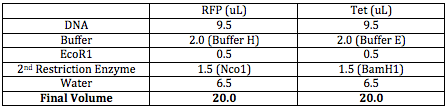
I double digested both RFP and Tet FML sequences. I used all 9.5 uL of my DNA. The following table outlines how much volume of each material I mixed together:
While I waited 3 hours for my digestion to take place, I prepared 6 tubes to make more liquid cultures of the RFP and Tet vectors to grow more receiving vectors. 3 tubes would be to make RFP cultures and the other 3 would be for the Tet cultures. For RFP, I used the part I715039 and for Tet, I used the part J31007.
Once the digestion took place, Dr. Campbell notified me that heat inactivation was capable for Nco1 but not BamH1. There were 3 options for me to take in order to clean the Tet FML, the protocols for all of which can be found on the Davidson Lab Protocols link:
a) [Clean and Concentrate DNA (after PCR, before digestion)]