Katie's Assignment
I am interested in exploring the genetic make up of enzymes that have been previously identified as being "salt dependent" for activity- such as citrate synthase, malic enzyme and aspartae transcarbamylase (which are all found in our species' genome).
What is citrate synthase and what is its role in our species?
What is malic enzyme (malate dehydrogenase) and what is its role in our species?
Why/How did I pick these three enzymes?
JGI Genes:
Citrate Synthase JGI: 2916896..2918032 (+) (1137bp). . . nucleotide sequence
Allosteric NADP-dependent Malic Enzyme 2055313..2057565 (+) (2253bp) . . . nucleotide sequence
Aspartate carbamoyltransferase regulatory subunit: 1503175..1503639 (-) (465bp). . . sequence
carbamoyltransferase: 1503636..1504550 (-) (915bp) . . . sequence
RAST Genes:
Citrate Synthase TCA Cycle. . . sequence . . . protein sequence
>fig|485914.5.peg.3029 [Halomicrobium mukohataei DSM 12286] [Citrate synthase (si) (EC 2.3.3.1)] MSDDLKQGLEGVLVTESELSKIDGDAGKLVYRGYTIEDLATGASFEEVLY LLWHGHLPNAAELDEFTDAMVEERHVDDDVMQTVEQLADADENPMAALRT AVSMLSSHDPDAETDPTDLDANLRKGRRITAKIPTVLAAFARFRDGQDAV EPREDLSHAANFLYMLNGEAPDEVLAETFDMALVLHADHGINASTFSAMV TASTLSDLHSAITSAIGTLKGSLHGGANQDVMEMLKEVDDAQQDPIDWVK TALDEGRRVSGFGHRVYNVKDPRAKILSQRSKELGEAAGSLKWYEMSTAI EDYLKAEKGLAPNVDFYSASTYYQMGIPIDIYTPIFAMSRVGGWTAHVLE QYENNRLIRPRARYVGPTDQTFVPLDER
Citrate Synthase Glyoxylate Synthesis . . . protein sequence
NADP-dependent malic enzyme. . . protein sequence
>fig|485914.5.peg.2113 [Halomicrobium mukohataei DSM 12286] [NADP-dependent malic enzyme (EC 1.1.1.40)] MGLDEDALDYHGRAPPGKIEIATTKPTNTQRDLSLAYSPGVAAPCEAIHE TPEDAFKYTARGNLVAVVSDGSAVLGLGDIGPEASKPVMEGKGVLFKRFA DIDVFDLELDTDDPDAMIEAVDAMGPTFGGINLEDIAAPACFEIERELRE RMDVPVFHDDQHGTAIISGAALLNAADIVDKELEEMEIVFSGAGASAIAS ARFYVSLGVRKENITMCDSSGIITADRVENDGLNRYKAEFASEGTGGDLA DALAGADAFVGLSVGGVVDEAMVRSMASEPIIFAMANPDPEIDYETAKAA RDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEINEEMKVAAARA LARLARQDVPDAVVKAYGDQPLQFGPEYIIPKPLDPRVLFEVTPAVAEAA MDSGAARKSIDLDDYVERLEARLGKSREMMRVVLNKAKSDPKRVVLAEGD DEKMIRAAYQLIEQGIAEPVLLGDRDRISAITDTLGLAFEPEIVDPDEGG LDEYADRLYELRQRKGVTRREADELVTDGNYLGSVMVEMGDADAMLTGLT HHYPSALRPPLQIVGTAPEAEYAAGVYMLTFRNRVVFCADTTVNTDPDAD VLTEVTRHTAELARRFNVEPRAAMLSYSNFGSVDSPSTRAPRRAAERLRE DPATDFPVDGEMQADTAVVEDILQGTYEFSELDDPANVLVFPSLEAGNIG YKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNMAGVAVVDAQDD
Aspartate carbamoyltransferase
>fig|485914.5.peg.1562 [Halomicrobium mukohataei DSM 12286] [Aspartate carbamoyltransferase (EC 2.1.3.2)] MRQDHIISAKQLSRRDIEAVLDRAAEIAADPSAYADRHEGSLLGLLFFEP STRTKMSFSAAMKRLGGDIVDMGTVESSSVKKGESLADTVRVVEGYADAL VLRHPSEGAAQMASEFVDAPLINAGDGAGQHPTQTLLDLYTIRENAGFDD LSIGIMGDLKYGRTVHSLAHALTVFDARQHFVSPESLQLPRSVRYDLHES GAEVREHTDLDDVLSELDVLYVTRIQKERFPDESEYHEVAGEYQIDAATI REHNEDLTVMHPLPRVDEIDHDVDELDGAQYFQQAHNGVPVRMALLDMVL EESR
Aspartate carbamoyltransferase regulatory chain
Main Tasks
1). Are the genes different between the annotation services?
The two citrate synthase genes annotated from RAST are the same gene. . . below are the nucleotide blast alignment results:


The citrate synthase gene from JGI matches the citrate synthase genes from RAST, nucleotide blast alignment dot plot is below:
The aspartate carbamoyltransferase regulatory gene sequences are a 100% match between the two annotation services.
The aspartate carbamoyltransferase gene sequences are the same between both annotation services as well.
The malic enzyme genes are 100% the same between the two annotation services as well.
I also checked to see if the aspartate carbamoyltransferase regulatory subunit was part of the aspartate carbamoyltransferase gene but it was not as confirmed by blastn with alignment and the nucleotide base regions where the gene occurs.
2). Look for other genes in our species' genome that are similar and may have been missed during annotation
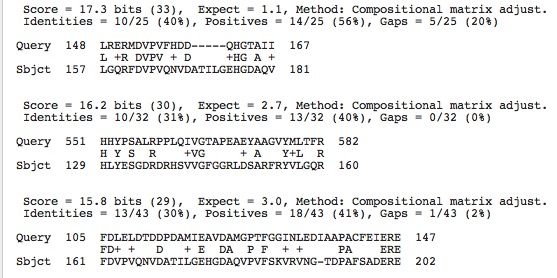
Citrate Synthase blasted against our genome using Genome Portal: Two hits that are small but maybe a conserved region between the three enzymes that makes them salt dependent. Note that the subject alignments do not fall within any of the other two genes we are studying and thus this correlated sequence within our species' genome is not from one of the other salt dependent enzyme genes.
Malic Enzyme blasted against our species' genome using Genome Portal: Three hits shown below:
Aspartate carbamoyltransferase blasted against our species' genome using Genome Portal, results are below: (no hits besides the original gene)
Aspartate carbamoyltransferase regulatory unit blasted against out species' genome using Genome Portal, results are below: (no other hits besides the original gene)
3). Note the pathways and systems that these genes play a role in
4). Look at the sequences from the halophile studied in the article as compared to these gene sequences
Lanyi in his paper, "Salt- Dependent Properties of Proteins from Extremely Halophilic Bacteria ," explores multiple enzymes that require salt to function properly. The three I have choosen to study: citrate synthase, malic enzyme and asparatate transcarbamylase, were isolated from H. cutirubrum. I had difficulty finding H. cutirubrum sequences in NCBI so I did some background research and discovered that H. cutirubrum is a specific strain of the H. salinarium species. According to Ventoso and Oren, there is no difference between this strain and the H. salinarium species.
The H. salinarium genome webpage outlines the three genes and below are the gene sequences:
Citrate Synthase . . . Protein Sequence and Information about the Protein
Found two possible sequences in H. salinarium's genome for malic enzyme: 1). malate dehydrogenase and 2).malate dehydrogenase (oxaloacetate decarboxylating) / phosphate acetyltransferase:
Malate Dehydrogenase (malic enzyme) . . .Protein Sequence and Information about the Protein
Malate Dehydrogenase (oxaloacetate decarboxylating) / phosphate acetyltransferase. . .Protein Sequence and Information about the Protein
The second malic enzyme sequence is most similar to our species' in length but both comparisons are shown below.
Aspartate Carbamoyltransferase . . . Protein Sequence and Information about Protein
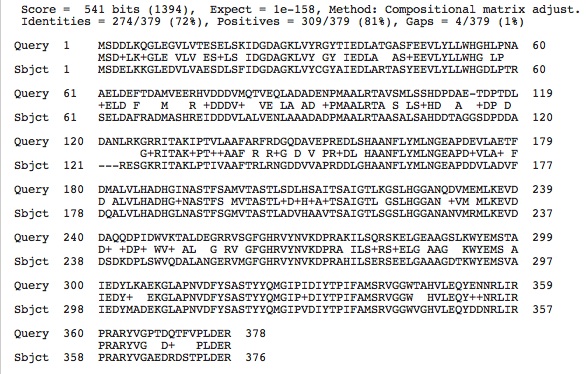
Comparison of Each Enzyme's Protein Sequences using blastp and clustalW. . .
Citrate Synthase Sequences:
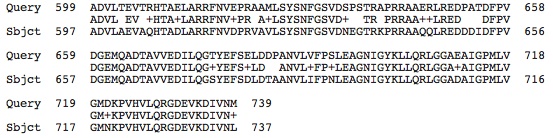
Malic Enzyme Sequences: (Query=our species' malic enzyme protein sequence and Subject= H. salinarium malic enzyme sequence) Malate Dehydrogenase:
Malate Dehydrogenase (oxaloacetate decarboxylating) / phosphate acetyltransferase:
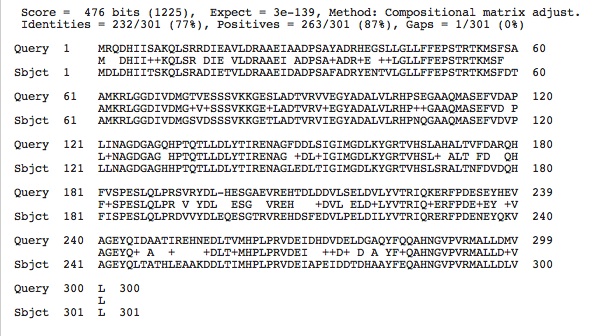
Aspartate Carbamoyltransferase Sequences: (Query= our species' and Subject=H. salinarium aspartate carbamoyltranserase enzyme protein sequence)
The high conservation between the Aspartate Carbamoyltransferase and Citrate Synthase between our species' enzyme protein sequences and that of the halophile studied may prove to show that these enzymes in our species are also dependent on high salt concentrations.
5). Find these genes in the other 8 genomes of halophiles that have been annotated and the closely related halophiles as indicated by Dr. C's phylogenetic tree
Citrate Synthase:
Haloarcula californiae ATCC 33799:
MSDDLKKGLEGVIVAESELSVIDGDAGKLVYRGYTIEDLAKGASY
EEVLYLLWHGHLPNRDELSEFKQAMVDARGVDDDVISTVRQLAEADENPMAALRTAVSM
LSAFDPAPEDAEPTDEMVNLETGRRITAKIPTIIAAFTRIRDGKEPVEPHDDLDHAANF
LYMLNGEEPDDVLADVFDQALVLHADHGLNASTFSAITTASTLSDVHSAVTSAIGTLKG
PLHGGANQDVMEMLKEVDDAESDPLDWVKNALDEGRRVSGFGHRVYNVKDPRAKILGER
SKELGEAAGTLKWYEMSTTIEDYLMEEKGLAPNVDFYSASTYYQMGIPIDIYTPIFAMS
RVGGWVAHVFEYIEDNRLIRPRARYVGKNTDETEFVPLDER
Haloarcula sinaiiensis ATCC 33800
MSDDLKKGLEGVIVAESELSVIDGDAGKLVYRGYTIEDLAKGASY
EEVLYLLWHGHLPNRDELSEFKQAMVDARGVDDDVISTVRQLAEADENPMAALRTAVSM
LSAFDPAPEDAEPTDEMVNLETGRRITAKIPTIIAAFTRIRDGKVPVEPRDDLDHAANF
LYMLNGEEPDDVLADVFDQALVLHADHGLNASTFSAITTASTLSDVHSAVTSAIGTLKG
PLHGGANQDVMEMLKEVDDAESDPLDWVKNALDEGRRVSGFGHRVYNVKDPRAKILGER
SKELGEAAGTLKWYEMSTTIEDYLMEEKGLAPNVDFYSASTYYQMGIPIDIYTPIFAMS
RVGGWVAHVFEYIEDNRLIRPRARYVGKNTDETEFVPLDER
Haloarcula vallismortis ATCC 29715
MSDDLKKGLEGVIVAESELSVIDGDAGKLVYRGYTIEDLAKGASY
EEVLYLLWHGHLPNRDELSEFKQAMVEARDVDDDVISTVRQLAEADENPMAALRTAVSM
LSAFDPAPEDAEPTDETVNLETGRRITAKIPTIIAAFTRIRDGKEPVEPRDDLDHAANF
LYMLNGEAPDDVLADVFDQALVLHADHGLNASTFSAITTASTLSDVHSAVTSAIGTLKG
PLHGGANQDVMEMLKEVDDAESDPLEWVQNALDEGRRVSGFGHRVYNVKDPRAKILGER
SKELGEAAGTLKWYEMSTTIEDYLMEEKGLAPNVDFYSASTYYQMGIPIDIYTPIFAMS
RVGGWVAHVFEYIEDNRLIRPRARYVGKNPDETTFVPLDER
Haloferax denitrificans ATCC 35960
MSGELKRGLEGVLVTESKLSFIDGDAGQLVYCGYDIEDLARDASY
EEVLYLLWHGELPTREELDAFSDELAAHRGLDDGVLDVARELAEQDESPMAALRTLVSA
MSAYDENADFEDVTDREVNLEKAKRITAKMPSVLAAYARFRRGDDYVEPDESLNHAANF
LYMLNGEEPNEVLAETFDMALVLHADHGLNASTFSAMVTSSTLSDLYSAVTSAIGTLSG
SLHGGANANVMRMLKDVDDSDMDPTEWVEDALDRGERVAGFGHRVYNVKDPRAKILGAK
SEALGEAAGDMKWYEMSVAIEEYIGEEKGLAPNVDFYSASTYYQMGIPIDLYTPIFAVS
RAGGWIAHVLEQYEDNRLIRPRARYTGEKGLEFPTVDER
Haloferax mediteranei ATCC 33500
MSGELKRGLEGVLVTESELSFIDGDAGQLIYRGYDIEDLARDASY
EEVLYLLWHGELPNRTQLDEFSDELAAHRDIGDGILDVARELAEQDESPMAALRTLVSA
MSAYDENADFEDVTDREVNLEKAKRITAKMPSVLAAYARFRRGDDYVAPNDDLNHAANF
LYMLNGEEPNEVLAETFDMALVLHADHGLNASTFSAMVTSSTLSDMYSAVTSAIGTLSG
SLHGGANANVMRMLKDVDDSDMDPVDWVEDALDRGERVAGFGHRVYNVKDPRAKILGQK
SEALGEAAGDMKWYEMSVAIEEYISEEKGLAPNVDFYSASTYYQMDIPIDLYTPIFAVS
RSGGWIAHILEQYDDNRLIRPRARYTGDKDLDFPTLDER
Haloferax mucosum ATCC BAA-1512 Haloferax sulfurifontis ATCC BAA-897 Haloferax volcanii ATCC 29605 Halorhabdus utahensis
Malic Enzyme:
Haloarcula californiae ATCC 33799
MGLDEDALEYHRSKPPGKIEIATTKPTNTQRDLSLAYSPGVAAPC
EEIDKDPEKAFEYTAKGNLVGVVSDGSAVLGLGDIGPEAGKPVMEGKGVLFKRFADIDV
FDVELDTDNAEAMIQTVEAMEPTFGGINLEDIAAPECFEVERRLSEKLDIPVFHDDQHG
TAIISGAALVNAADIADKELDELEVVFSGAGASAIASAKFYVSLGVSKDNITMCDSSGI
ITEERANHEELNRFKQEFARDIPEGGLADAMDGADVFVGLSVGGIVDQEMVRSMASDPI
IFAMANPDPEIAYEDAKAARDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EEMKVAAARALADLARQDVPDEVVKAYGDQPLQFGPDYIIPKPLDPRVLFEVTPAVAEA
AIESGAARTELDTAAYVEELEARLGKSREMMRVVLNKAKSDPQRVVLAEGHDEKMIRAA
YQLVEQGIAEPILIGDADRIESTRRKFGLEFDPVVVDPETADVADYADRLYELRQRKGV
TRREADELIRDGNYLGSVMVEMGDADAMLTGLTHHYPSALRPPLQVIGTADDADYAAGV
YMLTFKNRVIFCADTTVNQDPDTDVLEEVTRHTGELARRFNVEPRAAMLSYSNFGSVDN
LGTKKIRRAVSRLQDDDRVDFPVDGEMQADTAVVEDILQDTYEFSELDDPANVLVFPNL
EAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAGVAVVDAQ
Haloarcula sinaiiensis ATCC 33800
MGLDEDALEYHRSKPPGKIEIATTKPTNTQRDLSLAYSPGVAAPC
EEIDKDPEKAFEYTAKGNLVGVVSDGSAVLGLGDIGPEAGKPVMEGKGVLFKRFADIDV
FDVELDTDNAEAMIQTVEAMEPTFGGINLEDIAAPECFEVERRLSEKLDIPVFHDDQHG
TAIISGAALVNAADIADKELDELEVVFSGAGASAIASAKFYVSLGVSKDNITMCDSSGI
ITEERANHEELNRFKQEFARDIPEGGLADAMDGADVFVGLSVGGIVDQEMVRSMASDPI
IFAMANPDPEIAYEDAKAARDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EEMKVAAARALADLARQDVPDEVVKAYGDQPLQFGPDYIIPKPLDPRVLFEVTPAVAEA
AIESGAARTELDTAAYVEELEARLGKSREMMRVVLNKAKSDPQRVVLAEGHDEKMIRAA
YQLVEQGIAEPILIGDADRIESTRRKFGLEFDPVVVDPETADVADYADRLYELRQRKGV
TRREADELIRDGNYLGSVMVEMGDADAMLTGLTHHYPSALRPPLQVIGTADDADYAAGV
YMLTFKNRVIFCADTTVNQDPDTDVLEEVTRHTGELARRFNVEPRAAMLSYSNFGSVDN
LGTKKIRRAVSRLQDDDRVDFPVDGEMQADTAVVEDILQDTYEFSELDDPANVLVFPNL
EAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAGVAVVDAQQE
Haloarcula vallismortis ATCC 29715
MGLDEDALEYHRSKPPGKIEIATTKPTNTQRDLSLAYSPGVAAPC
NEIDEDPERAFEYTAKGNLVGVVSDGSAVLGLGDIGPEAGKPVMEGKGVLFKRFADIDV
FDVELDTDDAEAMIQTVAAMEPTFGGINLEDIAAPECFEVERRLSEKLDIPVFHDDQHG
TAIISGAALVNAADIAGKELEDLEVVFSGAGASAIASAKFYVSLGVSKDNITMCDSSGI
ITEERATHEDLNRFKQEFARDIPEGGLADAMDGADVFVGLSVGGIVDQEMVRSMASDPI
IFAMANPDPEIAYEDAKSARDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EEMKVAAARALADLARQDVPDEVVKAYGDQPLQFGPDYIIPKPLDPRVLFEVTPAVAEA
AIESGAARTEIDTDAYVEQLEARLGKSREMMRVVLNKAKSDPQRVVLAEGHDEKMIRAA
YQLVEQGIAEPVLIGDADQIESTRRKFGLEFDPVVVDPETADVADYADRLYEIRQRKGI
TRREAEELVRDGNYLGSVMVEMGDADAMLTGLTHHYPSALRPPLQVIGTADDADYAAGV
YMLTFKNRVIFCADTTVNQAPDADVLEEVTRHTGELARRFNVEPRAAMLSYSNFGSVDN
PGTKKIRRAVSRLQDDDRVDFPVDGEMQADTAVVEDILQDTYEFSELDEPANVLVFPNL
EAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAGVAVVDAQQE
Haloferax denitrificans ATCC 35960
MGLDDDSREYHRQDPPGKIEIATTKPTNTQRDLSLAYSPGVAAPC
LDIADDEDAAYEYTAKGNLVGVVSNGSAVLGLGNIGAQASKPVMEGKGVLFKRFADIDV
FDVELDITNVDEFVAATKAMEPTFGGINLEDIKAPECFEIERQLREQMDIPVFHDDQHG
TAIISGAALLNAADVTDKDLDDLDIVFSGAGASALATARFYVSLGAKKENITMCDSSGI
ITESRVEEGDVNKYKAEFAQPVDEGSLSDAIEGADVFAGLSVGGIVSQEMVRSMADDPI
IFAMANPDPEITYEDAKDAREDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EAMKRAAAEALAELARQDVPDAVVKAYGDHPLQFGPDYVIPKPLDPRVLFEVAPAVAQA
AMTSGAARERLDMNAYRERLEARLGKSREMMRVVLNKAKSNPKRVALAEGEDEKMIRAA
YQMQEEGIAEPILVGKTTTILRKAEELGLDFDPTIANPRDGEWDHYVDRLYELRQRKGV
TKSEAEELVRRDSNYFASVMVEVGDADAMLTGLTHHYPSALRPPLQIIGTAADANYAAG
VYMMTFKNRVVFCADTTVNLDPDEEILAEITKHTADLARQFNVEPRAALLSYSNFGSVT
NEGTRKPRDAAALLQGDPEVDFPVDGEMQADTALVEDILEGTYDFAELDGPANVLVFPN
LEAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAATAVVDAQEEE
Haloferax mediteranei ATCC 33500
MTLEDDARDYHRRPPSGKIEIATTKPTNTQRDLSLAYSPGVAAPC
LDIADDETVAYDYTAKGNLVGVVSNGSAVLGLGDIGAQASKPVMEGKGVLFKRFADIDV
FDVELGLDDPESFVQAVAAMEPTFGGINLEDIKAPECFEIEAGLRDEMSIPVFHDDQHG
TAIISGAALLNAVDIADKDRSSLQVTFAGAGAAATATARFYVSLGIPRENITMCDIDGI
LSERRADAGDLNEYTEPFARGVDDGELEDAMEGADVFVGLSVGGIVSQDMVRSMADNPI
IFAMANPDPEITYEDAKNARDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EAMKRAAAEALANLARQDVPDAVVKAYGDEPLQFGPDYLIPKPLDQRVLYEVTPAVAEA
AMESGAARRERDLDAYREELEARLGKSREMMRVVLNKAKSDPKRVALAEGEDEKMIRAA
SQLVEDGIAEPILIGRTTEILRTAEELGLDFDPTIANPHDGEWNHYVDHLYEQRQRKGL
TRTEAAELVQQDSNHFASVMVDIGDADAMLTGLTHHYSSALRPPLQLIGTAEDATYAAG
VYMLTFRNRVIFCADATVNLDPDEEVLAEVTKHTAELARRFNVEPRAALLSYSDFGSVN
NAGTAKPQNAVKRLHDDPDVDFPVDGEMQADTALVEEMLTDTYDFTELDGPANVLVFPN
LEAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAGIAVVDAQE
MGLDDDSREYHRRDPPGKIEIATTKPTNTQRDLSLAYSPGVAAPC
RDIDEDEDAAYEYTAKGNLVGVVSNGTAVLGLGDIGSQASKPVMEGKGVLFKRFADIDV
FDVELDITDVDDFVAATKAMEPTFGGINLEDIKAPECFEIERQLRETMDIPVFHDDQHG
TAIISGAALLNAADVLGKDLEDLEIVFSGAGASALATARFYVSLGAKKENITMCDSSGI
ITESRVSAGDVNKYKAEFAQDVEEGSLAHAMEGADVFAGLSVGGIVSQDMVRSMADNPI
IFAMANPDPEITYEDAKNARDDTVIMATGRSDYPNQVNNVLGFPFIFRGALDVRATEIN
EAMKRAAAEALAELARQDVPDAVVKAYGDHPLQFGPDYLIPKPLDPRVLFEVAPAVAQA
AMTSGAARERLDMNEYRERLEARLGKSREMMRVVLNKAKSDPKRVALAEGSNEKMIRAA
YQMQEEGIAEPILVGDTTTILRKAEELGLEFDPTIANPHDGEWNHYVDHLYERRRRKGL
TRTEAAELVQQDSNYFASVMVSMDDADAMLTGLTHHYPSALRPPLQLIGTAEDANYAAG
VYMMTFKNRVVFCADTTVNLDPDEEILAEITKHTAELARRFNVEPRAALLSYSNFGSVR
NEGTAKPRDAARLLQNDPEVDFPVDGEMQADTALVEDILEGTYDFAELDDPANVLIFPN
LEAGNIGYKLLQRLGGAEAIGPMLVGMDKPVHVLQRGDEVKDIVNLAATAVVDAQQE
Haloferax mucosum ATCC BAA-1512 Haloferax sulfurifontis ATCC BAA-897 Haloferax volcanii ATCC 29605 Halorhabdus utahensis
Aspartate Carbamoyltransferase:
Haloarcula californiae ATCC 33799
MRHDHIISAKQLSRGDIETVLDHAADIAADPGAFADRHSDTLLGL
LFFEPSTRTKMSFTTAMKRLGGDIVDMGSVESSSVKKGESLADTVRVVEGYTDALVLRH
PMEGSAKMASEFVDVPLVNAGDGAGQHPTQTLLDLYTIRENAGFDDLTIGIMGDLKYGR
TVHSLAHALTTVDASQHFISPESLQLPRSVRYDLHEAGAGIREHTELDDILPELDVLYV
TRIQAERFPDESEYREVAGQYQIDGD
Haloarcula sinaiiensis ATCC 33800
MRHDHIISAKQLSRGDIETVLDHAADIAADPGAFADRHSDTLLGL
LFFEPSTRTKMSFTTAMKRLGGDIVDMGSVESSSVKKGESLADTVRVVEGYTDALVLRH
PMEGSAKMASEFVDVPLVNAGDGAGQHPTQTLLDLYTIRENAGFDDLTIGIMGDLKYGR
TVHSLAHALTTVDASQHFISPESLQLPRSVRYDLHEAGAGIREHTELDDILPELDVLYV
TRIQAERFPDESEYREVAGQYQIDGDTLAAAKDDLTVMHPLPRVDEIAHDVDETTHAQY
FQQAHNGVPVRMALLDLMLGGDQ
Haloarcula vallismortis ATCC 29715
MRHDHIISAKQLSRGDIETVLDHAADIAADPGAFANRHSDTLLGL
LFFEPSTRTKMSFTTAMKRLGGDIVDMGSVESSSVKKGESLADTVRVVEGYTDTLVLRH
PMEGSAKMASEFVDVPLVNAGDGAGQHPTQTLLDLYTIRENAGFEDLTIGIMGDLKYGR
TVHSLAHALTTVDARQHFISPESLQLPRSVRYDLHEAGAGIREHTDLDEILPDLDVLYV
TRIQAERFPDESEYREVAGQYQIDADTLAAAKDDLTVMHPLPRVDEIAHDVDETTHAQY
FQQAHNGVPVRMALLDLMLGGDQ
Haloferax denitrificans ATCC 35960
MRQDHLISASHLSREDIEAVLDRAADIDADPAAFRQRHAGKVLGL
CFFEPSTRTRMSFDSAMKRLGGQTVDMGPVESSSVKKGETLADTVRVVEGYADALVLRH
PSEGAATMAAEFVDVPLVNAGDGAGQHPSQTLLDLYTIRENAGLDDLTIGIMGDLKYGR
TVHSLAEALTNFDASQHFISPESLRLPRNVRYDLHASGAQVREHTELDEVLPELDVLYV
TRIQRERFPDENEYRKVAGQYQIDSETLDAASDDLTIMHPLPRVDEISPDIDDTDHATY
FEQAHNGIPVRMALLDILLSQDR
Haloferax mediteranei ATCC 33500
MRQDHLISAAHLSREDIEAVLDRAAEIDDDTAAFRQRHAGKVLGL
CFFEPSTRTRMSFDTAMKRLGGQTVDMGPVESSSVKKGETLADTVRVVEGYADALVLRH
PSEGAATMAAEFVDVPLVNAGDGAGQHPSQTLLDLYTIRENAGLDDLTIGIMGDLKYGR
TVHSLAEALTNFDASQHFISPESLRLPRNVRYDLHASGAQVKEHTELDEVLPELDVLYV
TRIQRERFPDENEYRKVAGQYQIDAETLKAASDDLTVMHPLPRVDEISPDIDDTDHATY
FEQAHNGIPVRMALLDILLSQADD
Haloferax mucosum ATCC BAA-1512 Haloferax sulfurifontis ATCC BAA-897 Haloferax volcanii ATCC 29605 Halorhabdus utahensis
6). Blast results of our species' genes: pick halophiles, bacteria, and eukarya to compare nucleotide sequence and protein sequence to (separate salt-loving and non-salt loving)
7). Look for differences in predicted protein structure and potential amino acid bias. (ClustalW)
8). Look for similarities between the genes for these 3 enzymes that are salt dependent.