Difference between revisions of "Logic Gates - Emma Garren"
(→Cellular Logic with Orthogonal Ribosomes. (Rackham and Chin, 2006)) |
(→Cellular Logic with Orthogonal Ribosomes. (Rackham and Chin, 2006)) |
||
| Line 71: | Line 71: | ||
Two-input logic gates were constructed based on the interactions between synthetic O-rRNA and O-mRNA. | Two-input logic gates were constructed based on the interactions between synthetic O-rRNA and O-mRNA. | ||
| − | *<b>Input:</b> | + | *<b>Input:</b> Orthogonal ribosomes (O-ribosomes) |
| − | *<b>Gate:</b> | + | *<b>Gate:</b> Orthogonal mRNAs (O-mRNAs) |
| − | *<b>Output:</b> | + | *<b>Output:</b> Fluorescence |
| − | + | O-mRNAs contain ribosome-binding sequences (RBSs) that do not direct the translation of downstream genes by endogenous ribosomes. | |
| − | + | O-ribosomes translate O-mRNA, but do not significantly translate any of the thousands of cellular transcripts bearing cellular RBS sequences. | |
| − | Three distinct O-ribosome-O-mRNA pairs were isolated, with molecular specificities for independent function. This was accomplished through [[Hunter Stone - Synthetic Biology Seminar|directed evolution]]: A library of new potential RBSs were placed upstream of a novel fusion of the genes encoding chloramphenicol acetyltransferase and uracil phosphoribosyltransferase | + | Three distinct O-ribosome-O-mRNA pairs were isolated, with molecular specificities for independent function. This was accomplished through [[Hunter Stone - Synthetic Biology Seminar|directed evolution]] in two steps: |
| + | #A library of new potential RBSs were placed upstream of a novel fusion of the genes encoding chloramphenicol acetyltransferase and uracil phosphoribosyltransferase, and screened against 5-fluorouracil to select for the O-mRNA sequences that were not translated by endogenous ribosomes. | ||
| + | #The selected O-mRNAs were combined with a library of mutated 16s rRNA sequences, and these cells were grown in the presence of chloramphenicol to screen for those in which the mutant ribosomes translated the O-mRNAs. | ||
[[Figure 1. O-ribosomes and Boolean logic.]] Description: | [[Figure 1. O-ribosomes and Boolean logic.]] Description: | ||
Revision as of 03:38, 20 November 2007
Contents
Logic Gates in Synthetic Biology
Background: Logic Gates and Truth Tables
A logic gate, the building block for digital circuits, is a computing unit that performs a logical operation on one or more inputs and produces a single output. Gates are identified by their function, and each type of logic gate can be represented with a distinctive symbol. Inputs to outputs are read from left to right. A small circle on the right indicates the inversion of the output. A double line on the left indicates that the function is "exclusive."
A truth table is a useful way to describe the behavior or function of a logic gate. Logic states for both inputs and outputs are designated with 0's and 1's. Click here to see the most common logic gates and their truth tables.
Logic gates can be placed in parallel or serial combinations in order to perform more complex functions. Here are some examples:
Logic Gates in Synthetic Biology
Synthetic biology applies many of the principles of engineering to the field of biology in order to create biological devices which can ultimately be integrated into increasingly complex systems. These principles include standardization of parts, modularity, abstraction, reliability, predictability, and uniformity (Adrianantoandro et al., 2006). The application of engineering principles to biology is complicated by the inability to predict the functions of even simple devices and modules within the cellular environment. Some of the confounding factors are gene expression noise, mutation, cell death, undefined and changing extracellular environments, and interactions with the cellular context (Adrianantoandro et al., 2006). While for digital logic, inputs are either on or off (1 or 0), biological logic is sometimes leads to intermediate induction levels (Voigt, 2006).
In synthetic biology, logic gates are created by engineering the biochemical reactions that regulate various cellular processes, such as transcription, translation, protein phosphorylation, allosteric regulation, ligand/receptor binding, and enzymatic reactions. Although the diversity of biochemical reactions can make it difficult to combine different devices, these logic gates can be used to build complex systems with functions that have many practical applications. Mathematical modeling is used to predict the dynamics of the signaling and regulatory networks resulting from the logic gates.
Biomolecular Logic Gates: In Vitro
In vitro studies have been used to design combinations of molecules that have emergent properties related to information processing--molecular computing devices. Both the inputs and outputs consist of molecular species, with the output being a biologically active molecule. The extent to which these devices will be used with the cellular context is unclear--however, they are bound to inspire new directions for research in synthetic biology, and have potential applications in biochemical sensing, pathway engineering, and medical diagnosis and treatment.
An autonomous molecular computer for logical control of gene expression. (Benenson et al., 2004)
Overview:
- Input: Specific combination mRNA levels, serving as a simple model of a disease state (cancer)
- Output: “Drug” (or drug repressor) in the form of a single-stranded DNA sequence with known anti-cancer activity
Sequence recognition is used to control enzyme catalysis of covalent bond formation and breakage.
This paper describes an in vitro system that recognizes a specific combination of mRNA levels as its inputs, and performs a logical operation that results in the production of a molecule that can affect gene expression. The input mRNA levels have been designed to mimic a simplistic version of gene expression modeling cancer, and the output is a drug-like ssDNA with known anticancer activity. Therefore, the molecular computer is analogous to "a computational version of 'diagnosis'" (424). The molecular computers consist of a double-stranded DNA sequence with unique 7-bp sequences that recognize the “input” RNA. Each mRNA indicator is processed one at a time. An “inactivation tag” results in the displacement of the transition molecule, which destroys the computation fragment. An “activation tag” results in activation of the transition molecule. There are two separate molecular computers working simultaneously—one that releases the drug (a specific single-stranded DNA sequence) upon “positive diagnosis,” and another that releases the drug suppressor in response to “negative diagnosis.” The drug (or drug repressor) is incorporated into the DNA fragment as an inactive loop, protected by the double-stranded recognition sequences. If all of the transitions are “positive,” the ultimate output is drug administration (release of ssDNA). If any of the steps results in a “negative” transition, the output is a drug repressor. The design is flexible in that any sufficiently long mRNA molecule can be used as an indicator, and any ssDNA or short RNA molecule (up to at least 21-bp) can be designed as the output. PCR was used to for the experimental demonstration of both diagnosis and drug administration, and it is shown that the amount of active drug increases with the confidence of positive diagnosis. Future work could test the effectiveness of using alternate inputs (such as proteins), alternate outputs (such as RNA interference), as well as testing the function of the molecular computer in vivo.
Figure 1. Logical design of the molecular computer. Description:
Figure 2. Operation of the molecular computer. Description:
Enzyme-Free Nucleic Acid Logic Circuits. (Seelig et al., 2006)
Overview:
- Input:
- Output:
Nucleic acid devices are simplified by the predicability of base pairing. Although previous research has engineered nucleic acid logic switches based on hybridization and conformational changes in vivo, and this paper designs chemical logic gates that are capable of being combined into large, reliable circuits. These logic gates embody the following digital design principles: logic, cascading, restoration, fan-out, and modularity.
Benefits of this approach: both inputs and outputs are in the same form, which makes cascading possible (the output for one gate serves as the input for another gate.
Figure 1: Two-input AND gate. Description:
Cellular Logic Gates: In Vivo
Various types of molecular logic gates allow for the design of synthetic gene circuits... Modularity...
Synthetic Oscillators and Switches
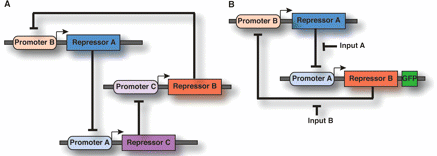 (Figure from Drubin et al., 2007. Permission pending.)
(Figure from Drubin et al., 2007. Permission pending.)
A. Repressilator (Elowitz and Leibler, 2000).
B. Toggle switch (Gardner et al., 2000). The toggle switch is also a form of cellular memory.
Environmental signal integration by a modular AND gate. (Anderson et al., 2007)
Modular AND gate Quorum-sensing as input (cellular communication)
Circuits engineered in the context of multicellular systems... Two different cell types in a bacterial population (Basu et al. 2004 and 2005, described in "Designing biological systems" review paper)
BioLogic Gates Enable Transcription Control in Mammalian Cells. (Kramer et al., 2004)
Cellular Logic with Orthogonal Ribosomes. (Rackham and Chin, 2006)
Overview:
Two-input logic gates were constructed based on the interactions between synthetic O-rRNA and O-mRNA.
- Input: Orthogonal ribosomes (O-ribosomes)
- Gate: Orthogonal mRNAs (O-mRNAs)
- Output: Fluorescence
O-mRNAs contain ribosome-binding sequences (RBSs) that do not direct the translation of downstream genes by endogenous ribosomes.
O-ribosomes translate O-mRNA, but do not significantly translate any of the thousands of cellular transcripts bearing cellular RBS sequences.
Three distinct O-ribosome-O-mRNA pairs were isolated, with molecular specificities for independent function. This was accomplished through directed evolution in two steps:
- A library of new potential RBSs were placed upstream of a novel fusion of the genes encoding chloramphenicol acetyltransferase and uracil phosphoribosyltransferase, and screened against 5-fluorouracil to select for the O-mRNA sequences that were not translated by endogenous ribosomes.
- The selected O-mRNAs were combined with a library of mutated 16s rRNA sequences, and these cells were grown in the presence of chloramphenicol to screen for those in which the mutant ribosomes translated the O-mRNAs.
Figure 1. O-ribosomes and Boolean logic. Description:
Applications and Future Directions
Synthetic gene circuits - use multiple simple input-output logic systems to design and build more complex circuits.
Medical applications - Cancer treatment - Increase specificity with which bacteria can sense an environment by combining multiple environmental inputs in logic gates. Autonomous biomolecular computing devices - use for molecular-level diagnostics and treatment
References
- Anderson, J. C., Voigt, C. A., Arkin, A. P. (2007). Environmental signal integration by a modular AND gate. Mol Syst Biol. 3:133. Abstract
- Andrianantoandro, E., Subhayu, B., Karig, D. K., Weiss, R. (2006). Synthetic biology: new engineering rules for an emerging discipline. Mol Syst. Biol. 2:2006.0028. Abstract
- Ashkenasy, G., Ghadiri, M. R. (2004). Boolean Logic Functions of a Synthetic Peptide Network. J Am Chem Soc. 126(36):11140-1. Abstract
- Baron, R., Lioubashevski, O., Katz, E., Niazov, T., Willner, I. (2006). Two coupled enzymes perform in parallel the “AND” and “InhibAND” logic gate operations. Org Biomol Chem. 4(6): 989-91. Abstract
- Baron, R., Lioubashevski, O., Katz, E., Niazov, T., Willner, I. (2006). Logic gates and elementary computing by enzymes. J Phys Chem A. 110(27):8548-53. Abstract
- Benenson, Y., Binyamin, G., Ben-Dor, U., Adar, R., Shapiro, E. (2004). An autonomous molecular computer for logical control of gene expression. Nature. 429:423-429. Abstract
- Boczko, E., Gedeon, T., Mischaikow, K. (2007). Dynamics of a simple regulatory switch. J Math Biol. 55(5-6):679-719. Abstract
- Chen, X., Wang, Y., Liu, Q., Zhang, Z., Fan, C., He, L. (2006). Construction of molecular logic gates with a DNA-cleaving deoxyribozyme. Angew Chem Int Ed Engl. 45(11):1759-62. Abstract
- Davidson, E.A., Ellington, A.D. (2007). Synthetic RNA circuits. Nat Chem Biol. 3(1):23-8. Abstract
- Dueber, J.E., Mirsky, E.A., Lim, W.A. (2007). Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat Biotechnol. 25(6):660-662. Abstract
- Drubin, D. A., Way, J. C., Silver, P. A. (2007). Designing biological systems. Genes Dev. 21(3):242-54. Abstract
- Elowitz, M. B., & Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature. 403(6767):335-8. Abstract
- Farfel, J., Stefanovic, D. (2005). Towards practical biomolecular computers using microfluidic deoxyribozyme logic gate networks. University of New Mexico.
- Frezza, B.M., Cockroft, S. L., Ghadiri, M.R. (2007). Modular Multi-level Circuits from Immobilized DNA-Based Logic Gates. J Am Chem Soc. (Epub ahead of print) Abstract
- Gardner, T.S., Cantor, C.R., Collins, J.J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature. 403(6767):338-42. Abstract
- Heinemann, M., Panke, S. (2006). Synthetic biology—putting engineering into biology. Bioinformatics. 22(22):2790-9. Abstract
- Kaznessis, Y. N. (2007). Models for synthetic biology. BMC Syst Biol. 1(1):47. Abstract
- Kramer, B. P., Fischer, C., Fussenegger, M. (2004). BioLogic Gates Enable Transcription Control in Mammalian Cells. Biotechnol Bioeng. 87(4):478-84. Abstract
- Lederman, H., Macdonald, J., Stefanovic, D., Stojanovic, M. N. (2006). Deoxyribozyme-based three-input logic gates and construction of a molecular full adder. Biochemistry. 45(4):1194-9. Abstract
- Narayanaswamy, R., Ellington, A.D. (2006). Engineering RNA-based circuits. Handb Exp Pharmacol. (173):423-45. Abstract
- Rackham, O., Chin, J. W. (2005). Cellular logic with orthogonal ribosomes. JACS 1227:17584-85. Abstract
- Rackham, O., Chin, J.W. (2006) Synthesizing cellular networks from evolved ribosome-mRNA pairs. Biochem Soc Trans. 34(2):328-9. Abstract
- Sayut, D.J., Kambam, P.K., Sun, L. (2007). Engineering and applications of genetic circuits. Mol Biosyst. 3(12):835-840. Abstract
- Seelig G., Soloveichik, D., Zhang, D. Y., Winfree, E., (2006). Enzyme-free nucleic acid logic circuits. Science. 314(5805): 1585-8. Abstract
- Stojanovic, M. N., Semova, S., Kolpashchikov, D., Macdonald, J., Morgan, C., Stefanovic, D. (2005). Deoxyribozyme-based ligase logic gates and their initial circuits. J Am Chem Soc. 127(19):6914-5. Abstract
- Voigt, C. A. (2006). Genetic parts to program bacteria. Curr Opin Biotechnol. 17:548-557. Abstract
- Wall, M. E., Hlavacek, W. S., Savageau, M. A. (2004). Design of gene circuits: lessons from bacteria. Nat Rev Genet. 5(1):34-42. Abstract
- Yan, H., Feng, L., LaBean, T.H., Reif, J.H. (2003). Parallel Molecular Computations of Pairwise Exclusive-Or (XOR) Using DNA "String Tile" Self-Assembly. J Am Chem Soc. 125:14246-14247. Abstract
- Yoshida, W., Yokobayashi, Y. (2007). Photon Boolean logic gates based on DNA aptamers. Chem Commun (Camb). (2):195-7. Abstract