Difference between revisions of "Mastering the Art of NCBI: It's a BLAST"
Clcarcelen (talk | contribs) |
Clcarcelen (talk | contribs) |
||
| Line 32: | Line 32: | ||
The most widely used database is the Nucleotide Collection (nr/nt) since it encompasses a broad range of nucleotide sequences across all domains, however you may choose to search another, depending on your research. | The most widely used database is the Nucleotide Collection (nr/nt) since it encompasses a broad range of nucleotide sequences across all domains, however you may choose to search another, depending on your research. | ||
| + | |||
| + | You may wish to restrict your search hits to only those found in certain organisms, or to exclude those found in a certain organism. You may do so by entering the common name, the binomial name or the taxonomic identification. | ||
| + | |||
| + | [[Image:Choose Organism.png]] | ||
| + | |||
| + | You have the option to further narrow your search using Entrez Query which limits to . This tool uses special and [http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml#entrez_query specific syntax] described on the NCBI website. This function is a specialized measure for narrowing search results, but it is only optional since the methods already described provide good results. | ||
| + | |||
Created by Claudia M. Carcelen, 2009. | Created by Claudia M. Carcelen, 2009. | ||
Revision as of 04:48, 8 October 2009
The National Center for Biotechnology Information (NCBI) is an organization founded in 1988 as a national resource available to the public for access to molecular biology information. NCBI creates numerous databases, online tools and research software programs to analyze genomes. The Basic Local Alignment Search Tool (BLAST) is an online tool designed to enable users to rapidly search through nucleotide and protein databases.While the website is designed for both novice and veteran users, the task of mastering the tool and the art can be daunting. This website is designed to provide a step-by-step process of how to use BLAST and interpret your results.
How to use Basic BLAST:
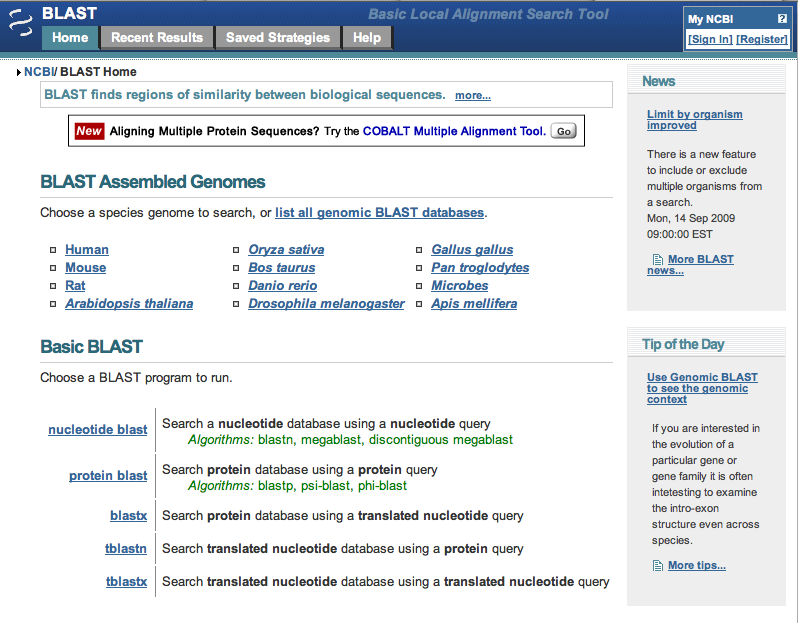
When you get to the main page, you may notice you have a number of options to choose from:
To search for matching nucleotide sequences in the database, choose: ![]()
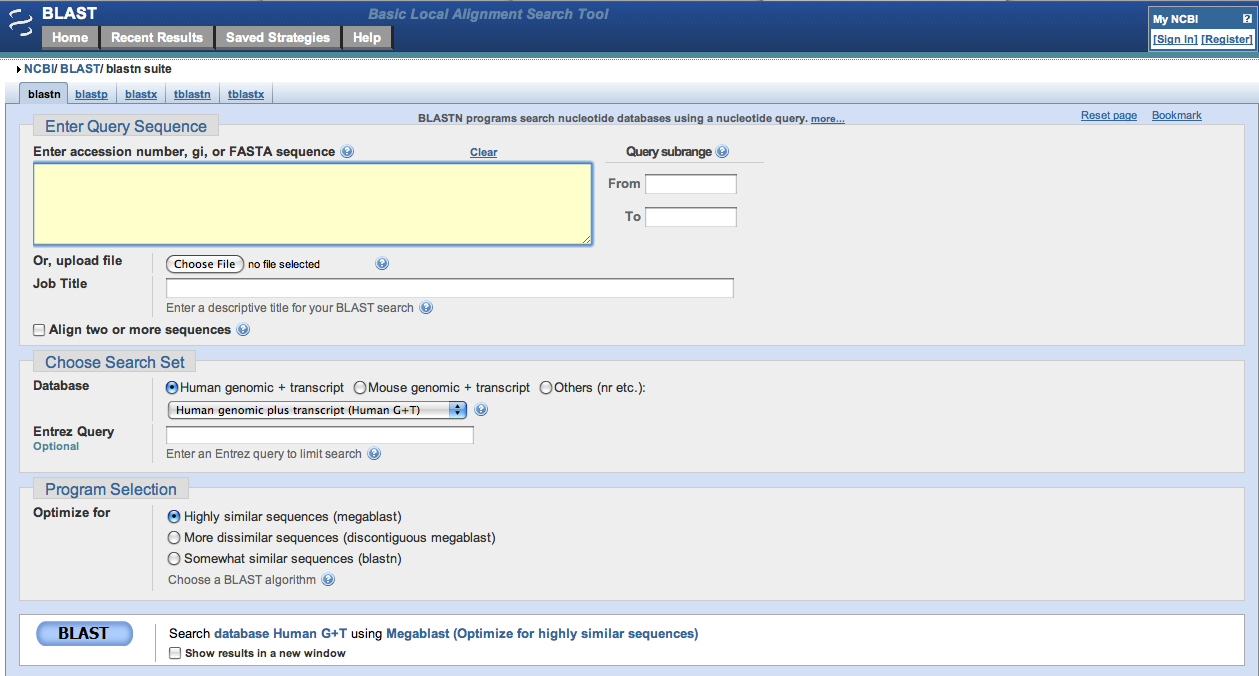
This link will take you to the page shown below.
In the entry box below Enter Query Sequence there are three possible methods of entry for your search. The first is bare sequence, which refers to simply to the nucleotide sequence (ATCG, etc.) you wish to search for.
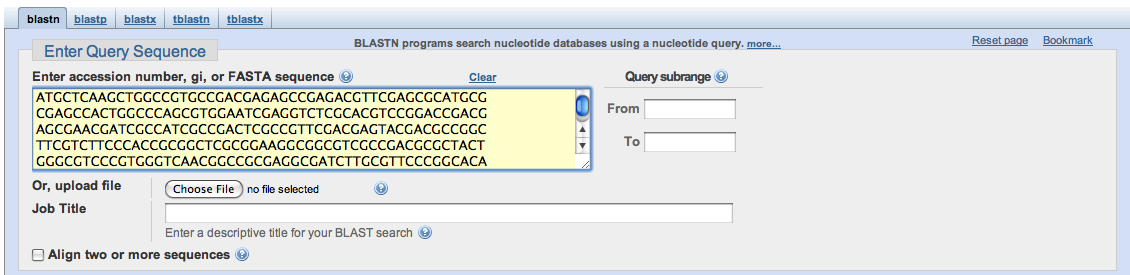
The second method uses FASTA format, shown below. This format requires the first line to be used as a descriptor, followed by a return and the nucleotide sequence. The descriptor can be found on the website where the gene sequence was obtained.
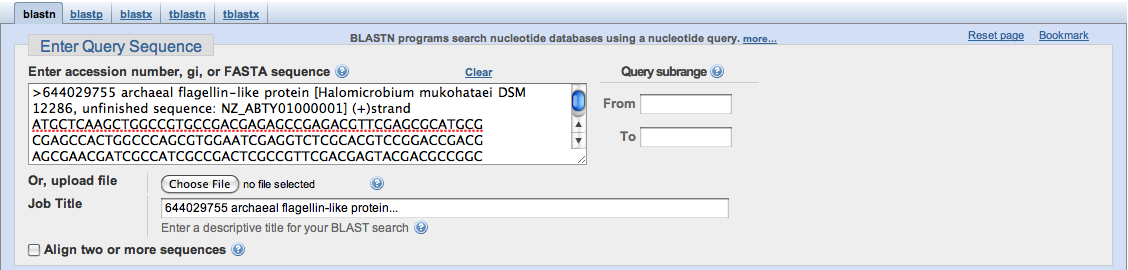
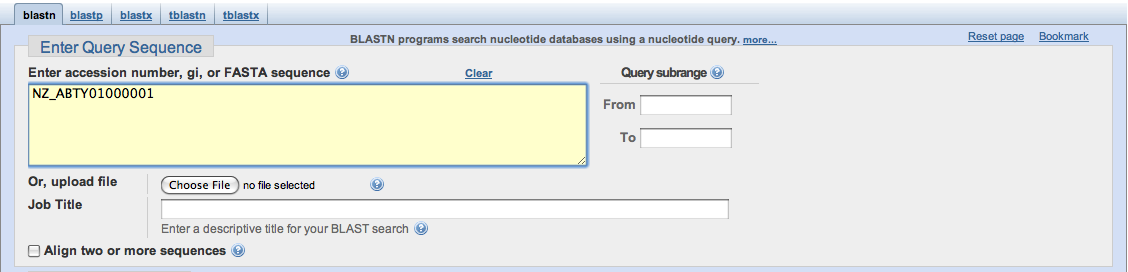
Finally, you may choose to use identifiers such as a gene's Accession Number as the query. It is important that there are no spaces in between letters or numbers, because they will be treated as separate sequences, or BLAST will fail to read them.
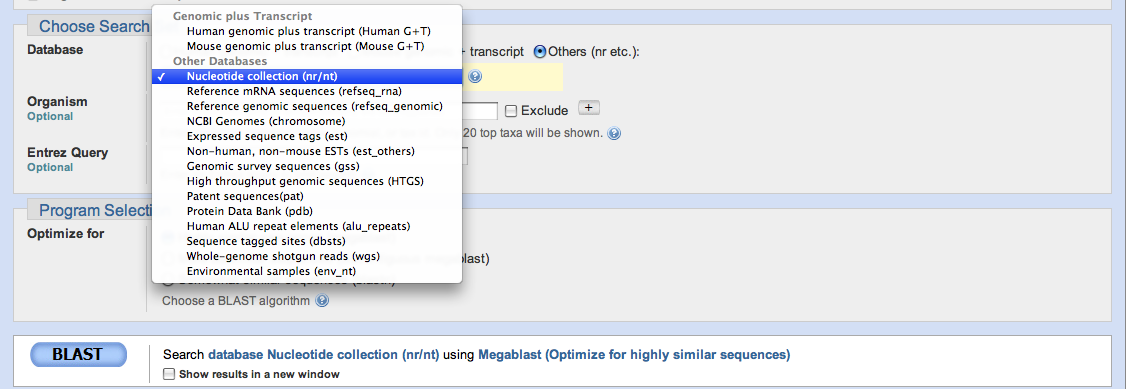
Once you have entered your query, you must choose which database you wish to search.
The most widely used database is the Nucleotide Collection (nr/nt) since it encompasses a broad range of nucleotide sequences across all domains, however you may choose to search another, depending on your research.
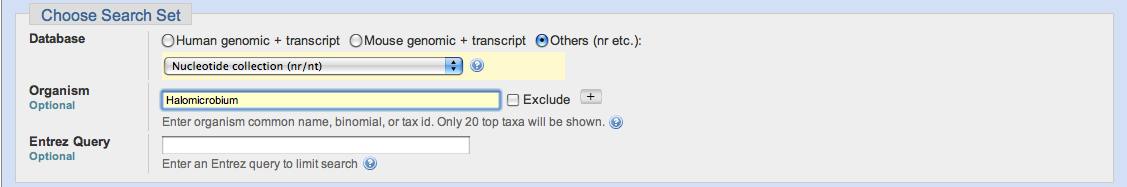
You may wish to restrict your search hits to only those found in certain organisms, or to exclude those found in a certain organism. You may do so by entering the common name, the binomial name or the taxonomic identification.
You have the option to further narrow your search using Entrez Query which limits to . This tool uses special and specific syntax described on the NCBI website. This function is a specialized measure for narrowing search results, but it is only optional since the methods already described provide good results.
Created by Claudia M. Carcelen, 2009.