Difference between revisions of "Protein Export"
MaCampbell (talk | contribs) (→Proteins Exported Completely) |
MaCampbell (talk | contribs) (→Proteins Exported Completely) |
||
| Line 34: | Line 34: | ||
*No TatAt in annotations | *No TatAt in annotations | ||
*No TatB in annotations | *No TatB in annotations | ||
| − | + | *No TorD in annotations | |
Tat proteins use protein domain of SRRXFLK can we find this? They transport fully folded proteins. Many halophiles have a lot of K+ in cytoplasm to counter balance extracellular salt. Their proteins have many negative amino acids that help keep the proteins and bacteria from “salting out” and may lead to rapid protein folding. Therefore, the Tet system may be more important than the Sec system. Figure from Sonja-Verena Albers, Zalán Szabó and Arnold J. M. Driessen. 2006. Nature Reviews: Microbiology. VOLUME 4. | Tat proteins use protein domain of SRRXFLK can we find this? They transport fully folded proteins. Many halophiles have a lot of K+ in cytoplasm to counter balance extracellular salt. Their proteins have many negative amino acids that help keep the proteins and bacteria from “salting out” and may lead to rapid protein folding. Therefore, the Tet system may be more important than the Sec system. Figure from Sonja-Verena Albers, Zalán Szabó and Arnold J. M. Driessen. 2006. Nature Reviews: Microbiology. VOLUME 4. | ||
Revision as of 16:15, 13 November 2008
Proteins On the Cell Surface
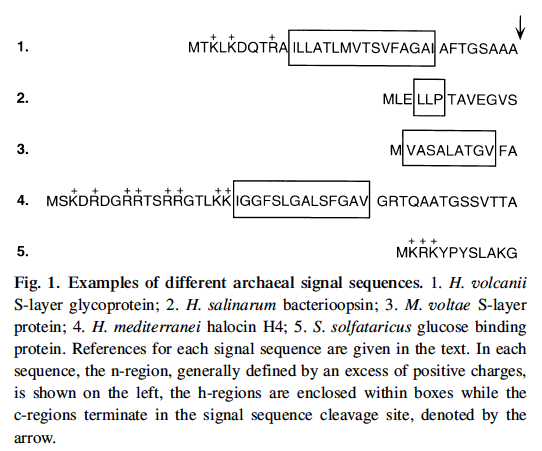
First, we should briefly consider proteins that reside in cell membrane. Like all other organisms, Halophile proteins begin with a signal sequence (20 - 30 residues long) that has many positive charges on it. We do not know enough to search for these in a reliable way. See five examples in the figure below from a review paper by Jerry Eichler, 2000. Archaeal protein translocation: Crossing membranes in the third domain of life. Eur. J. Biochem. 267: 3402-3412.
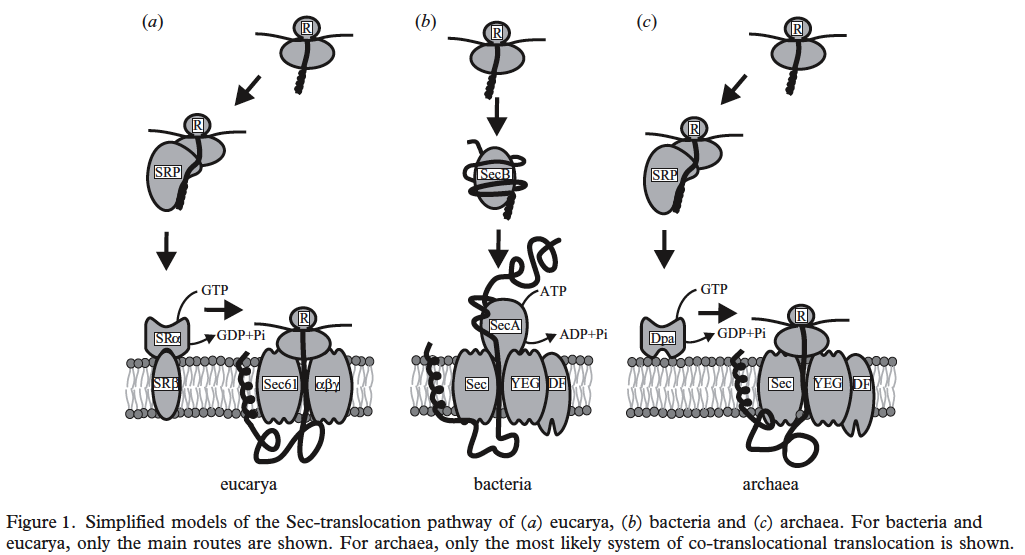
Three-domain comparison from Albert Bolhuis. 2004. The archaeal Sec-dependent protein translocation pathway. Phil. Trans. R. Soc. Lond. B. Vol. 359: 919–927.
Proteins Exported Completely
There are three main systems for protein export.
http://www.membranetransport.org/
1. Sec pathway using SecA, D, E, Y C F proteins.
- Our genome does not have any SecA annotations.
SPaseI and II cleave peptides off for export. Sec11a and sec11b have been found in halophiles.
YidC Oxa 1p
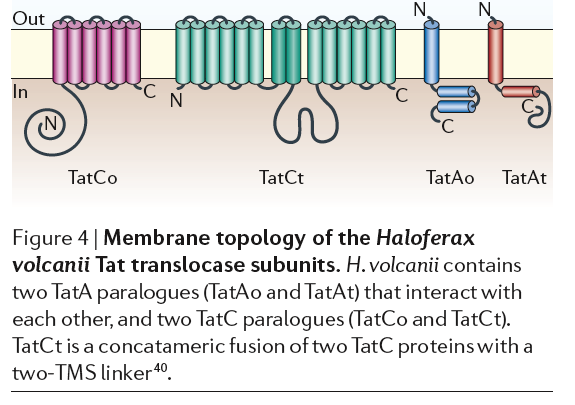
2. Tat pathway using TatA, TatCo, TatCt, TatAo TatAt, TatB, TorD and TorA are chaperones that bind to signal sequences bound for Tat protein export.
Our Organism has these genes:
- ORF01012 sec-independent protein translocase component TatA 1
- No TatCo in annotations
- No TatCt in annotations
- No TatAo in annotations
- No TatAt in annotations
- No TatB in annotations
- No TorD in annotations
Tat proteins use protein domain of SRRXFLK can we find this? They transport fully folded proteins. Many halophiles have a lot of K+ in cytoplasm to counter balance extracellular salt. Their proteins have many negative amino acids that help keep the proteins and bacteria from “salting out” and may lead to rapid protein folding. Therefore, the Tet system may be more important than the Sec system. Figure from Sonja-Verena Albers, Zalán Szabó and Arnold J. M. Driessen. 2006. Nature Reviews: Microbiology. VOLUME 4.
3. lipobox pathway for lipid-modified proteins, lipobox = [I/L/G/A][A/G/S]"cut_here"C is the target for SPaseII enzymes.
Type I – V secretion systems possible. I is ABC protein transporter
Flagella and pilus or pili proteins. PibD trims off cytoplasmic parts of flagellar proteins