Difference between revisions of "RRNA operon"
(→rRNA Operon) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ---- | |
| + | |||
| + | The rRNA operon encodes for the ribosomal RNA, which comprises the ribosomes. Three distinct rRNA sequences exist in the operon, encoding for the two subunits of the ribosome; the 5S and 23S sequences code for the large subunit, while the 16S sequence encodes for the small subunit. | ||
| + | |||
| + | The three sequences are coded in the following order, from 3' to 5': 16S, 23S, 5S. | ||
| − | ---- | + | The 3' end of the 16S RNA, or small subunit, binds to the 5' end of the messenger RNA during transcription, specifically to the [http://en.wikipedia.org/wiki/Shine-Dalgarno_sequence Shine-Dalgarno sequence]. This aligns the mRNA's start codon for transcription. The 16S rRNA itself contains an anti-Shine-Dalgarno sequence on its 3' end. |
| − | + | Along with a promoter, a termination sequence, and an operator, the three seperate segments coding for the different subunits of the ribosome all form a single operon. The specific promoter and termination sequence is not known, but a [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=ScaffoldGraph&page=scaffoldDna&scaffold_oid=2500575882&start_coord=2396557&end_coord=2397346 790-base-pair intergenetic region] exists upstream of the 16S coding region; the promoter sequence most likely resides there. | |
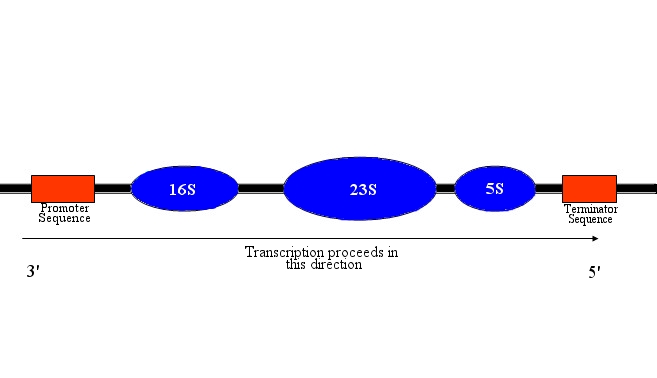
| + | [[Image:Diagram_operon.jpg]] | ||
| + | <p><small>Fig. 1. Graphical representation of the rRNA operon. Coding sequences for rRNA are grouped into three distinct clusters, with 16S representing the small ribosomal subunit and 23S and 5S forming the large subunit. The 16S cluster contains a Shine-Dalgarno sequence near its 3' end, to allow the small ribosomal unit to bond with mRNA. The operon begins with a promoter sequence and an operator sequence, currently unidentified, and transcription is halted by a terminator sequence, also currently unidentified.</small> | ||
| + | <br /> | ||
{| border="1" cellpadding="2" | {| border="1" cellpadding="2" | ||
| | | | ||
|- | |- | ||
| − | ! Gene Name !! Gene Coordinates !! Number of Base Pairs (Length) !! Score of Nearest | + | ! Gene Name !! Gene Coordinates !! Number of Base Pairs (Length) !! Closest Match on BLAST (Homolog) !! Score of Nearest Match !! E-value of Nearest Match !! Percent Identity to Nearest Match |
|- | |- | ||
! '''16S rRNA''' | ! '''16S rRNA''' | ||
| − | | 2397347..2398825 || 1479 bp || 1966 || 0.0e+00 || 92.12% | + | | 2397347..2398825 || 1479 bp || [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=638154504 Halobacterium sp. NRC - 1] || 1966 || 0.0e+00 || 92.12% |
|- | |- | ||
! '''Gap Bases''' | ! '''Gap Bases''' | ||
| − | | | + | | ''2398825..2399190'' || 365 || N/A || N/A || N/A || N/A |
|- | |- | ||
! '''23S rRNA''' | ! '''23S rRNA''' | ||
| − | | 2399190..2402100 || 2911 bp || 1899 || 0.0e+00 || 90.06% | + | | 2399190..2402100 || 2911 bp || [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=637000187 Natronomonas pharaonis DSM 2160] || 1899 || 0.0e+00 || 90.06% |
|- | |- | ||
! '''Gap Bases''' | ! '''Gap Bases''' | ||
| − | | | + | | ''2402100..2402216'' || 116 || N/A || N/A || N/A ||N/A |
|- | |- | ||
! '''5S rRNA''' | ! '''5S rRNA''' | ||
| − | | 2402216..2402338 || 123bp || 143 || 3.0e-36 || 90.00% | + | | 2402216..2402338 || 123bp || [http://imgweb.jgi-psf.org/cgi-bin/img_edu_v260/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=638154504 Halobacterium sp. NRC - 1] || 143 || 3.0e-36 || 90.00% |
|} | |} | ||
Latest revision as of 06:59, 2 September 2008
The rRNA operon encodes for the ribosomal RNA, which comprises the ribosomes. Three distinct rRNA sequences exist in the operon, encoding for the two subunits of the ribosome; the 5S and 23S sequences code for the large subunit, while the 16S sequence encodes for the small subunit.
The three sequences are coded in the following order, from 3' to 5': 16S, 23S, 5S.
The 3' end of the 16S RNA, or small subunit, binds to the 5' end of the messenger RNA during transcription, specifically to the Shine-Dalgarno sequence. This aligns the mRNA's start codon for transcription. The 16S rRNA itself contains an anti-Shine-Dalgarno sequence on its 3' end.
Along with a promoter, a termination sequence, and an operator, the three seperate segments coding for the different subunits of the ribosome all form a single operon. The specific promoter and termination sequence is not known, but a 790-base-pair intergenetic region exists upstream of the 16S coding region; the promoter sequence most likely resides there.
Fig. 1. Graphical representation of the rRNA operon. Coding sequences for rRNA are grouped into three distinct clusters, with 16S representing the small ribosomal subunit and 23S and 5S forming the large subunit. The 16S cluster contains a Shine-Dalgarno sequence near its 3' end, to allow the small ribosomal unit to bond with mRNA. The operon begins with a promoter sequence and an operator sequence, currently unidentified, and transcription is halted by a terminator sequence, also currently unidentified.
| Gene Name | Gene Coordinates | Number of Base Pairs (Length) | Closest Match on BLAST (Homolog) | Score of Nearest Match | E-value of Nearest Match | Percent Identity to Nearest Match |
|---|---|---|---|---|---|---|
| 16S rRNA | 2397347..2398825 | 1479 bp | Halobacterium sp. NRC - 1 | 1966 | 0.0e+00 | 92.12% |
| Gap Bases | 2398825..2399190 | 365 | N/A | N/A | N/A | N/A |
| 23S rRNA | 2399190..2402100 | 2911 bp | Natronomonas pharaonis DSM 2160 | 1899 | 0.0e+00 | 90.06% |
| Gap Bases | 2402100..2402216 | 116 | N/A | N/A | N/A | N/A |
| 5S rRNA | 2402216..2402338 | 123bp | Halobacterium sp. NRC - 1 | 143 | 3.0e-36 | 90.00% |