Riboregulators
After observing the many and varied naturally occuring post-transcriptional, regulatory RNA systems, Isaacs et al. designed and engineered a modular synthetic system where RNA turns on and off gene expression by controlling translation. The modularity of their system allows any gene to be regulated instead of only a specific gene to which the riboregulator is targeted.
Design
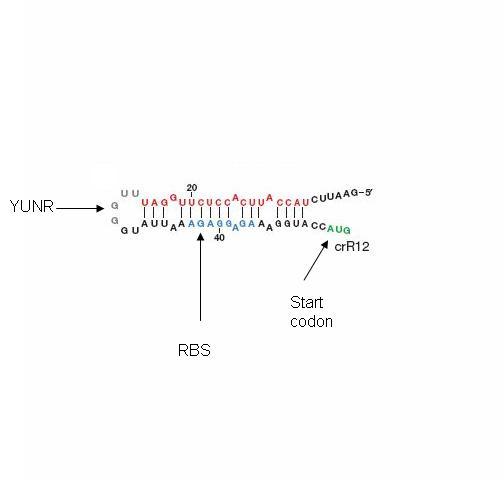
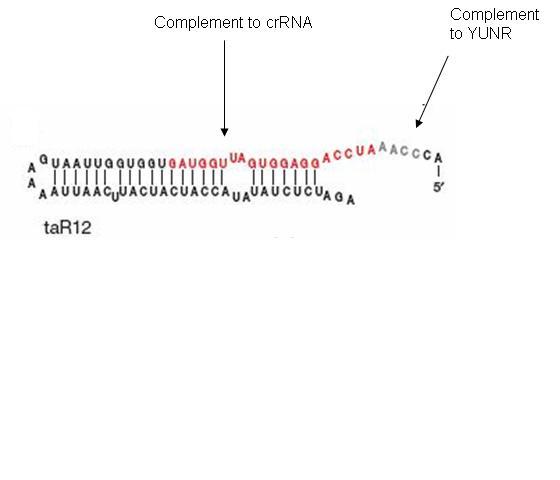
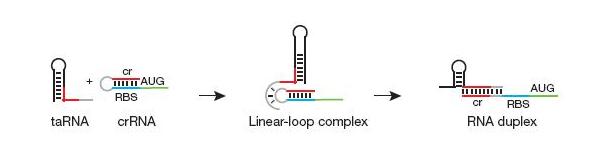
The design itself has two components: a short cis-repressed RNA sequence (crRNA) that is inserted upstream of the gene and a transactivating RNA sequence (taRNA) that targets the crRNA. The crRNA sequence contains two fundamental components: the complement of the ribosomal binding site (RBS) and a pyrimidine-uracil-nucleotide-purine (YUNR) sequence. When not interacting with the taRNA, the complement of the RBS binds to the RBS, causing the crRNA to loop and block the ribosome's access to the RBS. When the RBS is blocked, translation does not occur; the gene expression is off. The YUNR sequence has a complement on the taRNA. When the taRNA finds a crRNA, the interaction with the YUNR sequence begins pulling the crRNA off the RBS.
The taRNA is free floating and regulated by an inducible promoter. The inducible promoter allows the researcher to determine under what conditions gene expression will be turned on. Besides the complement to the YUNR sequence, the taRNA contains a section of high complementarity to section of the crRNA that folds over to cover the RBS, which allows the taRNA to keep the crRNA sequestered from the RBS.
To recap, gene expression is off when there is crRNA upstream of the gene but no taRNA is in the system. In the presence of taRNA, gene expression is turned back on.
From Concept to Wet Lab
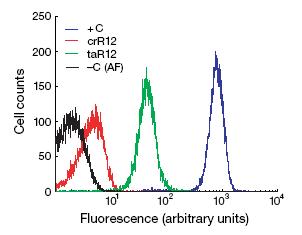
In wet lab, the crRNA sequence was placed in front of the GFP gene and introduced to cells. Using flow cytometry, Isaacs et al. measured fluorescence of cells that had just the GFP gene, the GFP gene with crRNA upstream, the GFP gene with crRNA upstream and taRNA, and no GFP at all. The crRNA decreased fluorescence to near basal levels. When taRNA was present in the system, fluorescence increased by approximately a power of ten. While the fluorescence did not equal fluorescence of cells with just GFP, the repression of gene expression with crRNA and return of expression with taRNA are strong enough to suggest the riboregulator system works.
These experiments were done with pBad and pLac controlling the production of taRNA. As the system worked when the taRNA was under control of either promoter and the crRNA can be inserted upstream of any gene, this system is considered modular.
Finally, riboregulators can control different genes in response to different stinuli by using different crRNA/taRNA pairs as the pairs are specific to eachother.