Riboswitches
Riboswitches are small sequences in mRNA that bind small molecules to regulate translation and occasionally transcription. Riboswitches occur naturally in both eukaryotes and prokaryotes. Desai and Gallivan hoped to find new synthetic riboswitches (riboswitches with new ligand specificities) by creating libraries of mutant riboswitches and using genetic selection to pick the functional ones of interest. Desai and Gallivan also employed riboswitches to screen for the presence of specific small molecules. In theory riboswitches are perfect because the number of aptamers already in existence and our capbaility to engineer new aptamers through rational design.
Design
Reviews of previous research showed that the theophylline aptamer worked in riboswitches in wheat germ, a eukaryotes, and Bacillus subtilis, a gram positive bacterium. Desai and Gallivan decided to translate the technology to gram negative bacteria. To do this they cloned the theophylline aptamer five base pairs upstream of the RBS for lacZ. The gene is controlled by a weak promoter and a weak RBS allowing for sensitivity to changes in translation because of the presence of theophylline. The construct was then transformed into E. coli. When theophylline is added, translation should be turned on again.
From Concept to Wet Lab
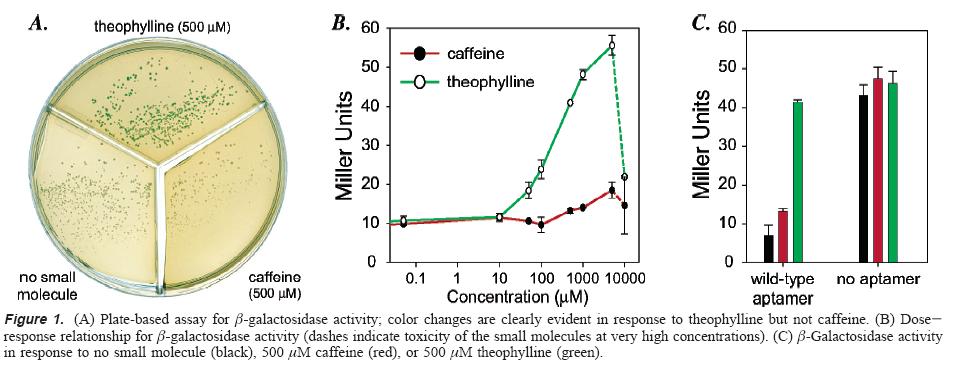
Simply using a visual check, a difference in color is visible between cells with the riboswitch but no theophylline or cells with the riboswitch and caffeine but no theophylline and cells with the riboswitch and theophylline. Quantification of the amount of beta-galactosidase showed that when theophylline was added the translation increased significantly above base-line. When no aptamer was cloned to make a riboswitch, beta-galactosidase did not vary significantly among the conditions of no ligand, caffeine, and theophylline.
To further support that the theophylline was interacting with the aptamer and not just increasing protein translation through another route. By introducing a single point mutation that in vitro decreases affinity for theophylline and increases affinity for 3-methylxanthine, the lab demonstrated that the translation in the presence of theophylline now was almost the same as translation without any small molecule while translation in the presence of 3-methylxanthine was significantly higher than base-line. These results suggest that the change in translation is controlled by the riboswitch and not some other mechanism.