Difference between revisions of "Semi-Synthetic DNA Shuffling and Doramectin"
(→The Experiment) |
(→Significance of DNA Shuffling) |
||
| Line 25: | Line 25: | ||
''S. avermitilis'' is a strain of bacteria which grows at very slow rates. Because of the slow growth of this bacterium, researchers reported a single round of directed evolution with ''S. avermitilis'' took an estimated 2-3 months. | ''S. avermitilis'' is a strain of bacteria which grows at very slow rates. Because of the slow growth of this bacterium, researchers reported a single round of directed evolution with ''S. avermitilis'' took an estimated 2-3 months. | ||
| − | == | + | ==Semi-synthetic DNA Shuffling== |
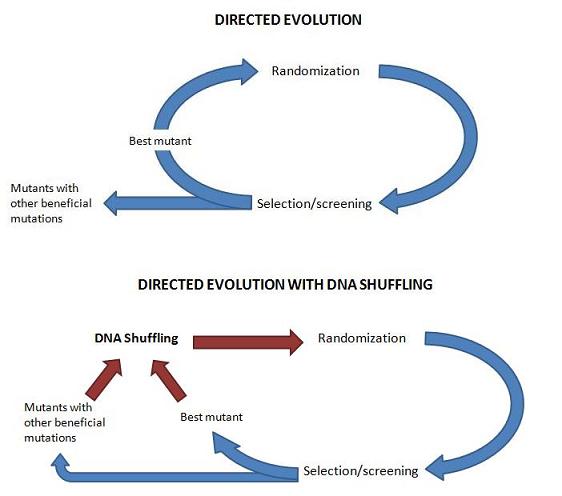
| − | During the process of genetic randomization in this experiments, the researchers used a technique termed DNA shuffling. This technique, sometimes | + | During the process of genetic randomization in this experiments, the researchers used a technique termed DNA shuffling. This technique, sometimes called sexual PCR, artificially recombining the genes of the best mutants at the end of each round of selection. |
| − | by selection | + | The first step of DNA shuffling involves digesting the gene of interest with DNas1 into fragments. PCR without primers is then conducted on these fragments. During primerless PCR, ''Taq'' polymerase is able to reassemble the whole gene from these fragments. Hopefully, during this fragmenting and reassembly of the gene of interest, different mutations proven beneficial by selection during directed evolution are recombined into a single gene (''Fig. 2a''). When these events occur, beneficial mutations are preserved from generation to generation of directed evolution that would normally be lost when only the best mutant is resubmitted to randomization (''Fig. 2b''). |
| − | |||
| − | |||
| − | |||
| − | + | [[Image:DNAshufflingtheorysmall.jpg]] | |
| + | |||
| + | |||
| − | |||
| − | + | The work by Stutzman-Engwall ''et al.'' represents a further development on the technique of DNA shuffling. They have termed their new method for shuffling "semi-synthetic DNA shuffling." This method follows the same protocol as DNA shuffling described above, except that all beneficial mutations of the ''aveC'' gene were stored as oligonucleotides. By this method, beneficial mutations can be continuously reintroduced by inserting these oligonucleotides in high molar concentrations to the DNA shuffling protocol. The researchers hypothesized this new development in DNA shuffling would prevent beneficial mutations from being lost in false negatives, speeding directed evolution ever further. | |
| − | + | The relationship between rounds of selection with semi-synthetic DNA shuffling, B2:B1 ratios in doramectin production, and number of mutations compared to wild-type ''aveC'' is compared in the following figure. | |
| − | + | ==Conclusion== | |
| − | '' | + | Semi-synthetic shuffling proved successful in quickly and efficiently improving the function of the ''aveC'' gene. |
==The Experiment== | ==The Experiment== | ||
Revision as of 03:27, 6 December 2007
Contents
Master
Background
Doramectin is a drug used to treat gastrointestinal roundworms, lungworms, eyeworms, grubs, and sucking lice in cattle. The drug is one of several avermectins, compounds used for the treatment of parasites in animals and river blindness in humans. According to the authors, “avermectin derivatives are the most widely used drugs in animal health and agriculture, with current worldwide sales exceeding 1 billion US dollars.”
Avermectins are produced by the soil-borne bacteria Streptomyces avermitilis. The subspecies Streptomyces is the largest genus of antibiotic-producing bacteri; erythromycin, neomycin, streptomycin, and tetracycline were all originally derived from species of Streptomyces.
The Goal
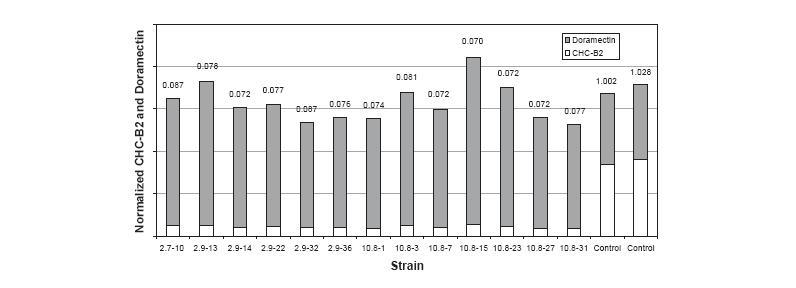
Researchers Stutzman-Engwall et al. began their experiment with a strain of S. avermitilis capable of producing doramectin from supplemented cyclohexancaroxylic acid. This strain produced doramectin in two forms: CHC-B1, the most useful form of doramectin, and CHC-B2, a related compound less effective as an antiparasital than CHC-B1. The ratio of uneffective CHC-B2 to effective CHC-B1 produced by this strain was 1:1.
The researchers were interested in creating a strain of S. avermitilis capable of producing higher yields of the B1 form of doramectin. The team had already identified that the gene aveC was responsible for the B2:B1 ratio. This knowledge led the team to conduct directed evolution upon the aveC gene.
The Experiment
The researchers conducted three rounds of directed evolution on the aveC gene. To generate a mutant library of the gene, the researchers used the mutagenic agent Mutazyme and semi-synthetic DNA shuffling (see following section). The mutant aveC genes were inserted into gene replacement vectors, which were then transformed into S. avermitilis cells. These cells were diluted into 96 well plates for high throughput culturing under conditions suitable for doramectin production. Selection of the best mutants was conducted by testing the doramectin output and B2:B1 ratios of each well (quantifiable with MS/MS) and then returning to culture to find this particular mutant. DNA isolated was isolated from these mutants and the aveC gene was amplified using PCR. The DNA of the best mutants from that generation were recombined using semi-synthetic shuffling, randomized, and resubmitted to selection.
The resulting strains of S. avermitilis after three rounds of directed evolution produced doramectin with a B2 to B1 ratio of 0.07:1 (Fig. 1). This ratio represented a significant decrease compared to the ratio of B2 to B1 in the found in the wild type strain of S. avermitilis (1:1).
The Problem
S. avermitilis is a strain of bacteria which grows at very slow rates. Because of the slow growth of this bacterium, researchers reported a single round of directed evolution with S. avermitilis took an estimated 2-3 months.
Semi-synthetic DNA Shuffling
During the process of genetic randomization in this experiments, the researchers used a technique termed DNA shuffling. This technique, sometimes called sexual PCR, artificially recombining the genes of the best mutants at the end of each round of selection.
The first step of DNA shuffling involves digesting the gene of interest with DNas1 into fragments. PCR without primers is then conducted on these fragments. During primerless PCR, Taq polymerase is able to reassemble the whole gene from these fragments. Hopefully, during this fragmenting and reassembly of the gene of interest, different mutations proven beneficial by selection during directed evolution are recombined into a single gene (Fig. 2a). When these events occur, beneficial mutations are preserved from generation to generation of directed evolution that would normally be lost when only the best mutant is resubmitted to randomization (Fig. 2b).
The work by Stutzman-Engwall et al. represents a further development on the technique of DNA shuffling. They have termed their new method for shuffling "semi-synthetic DNA shuffling." This method follows the same protocol as DNA shuffling described above, except that all beneficial mutations of the aveC gene were stored as oligonucleotides. By this method, beneficial mutations can be continuously reintroduced by inserting these oligonucleotides in high molar concentrations to the DNA shuffling protocol. The researchers hypothesized this new development in DNA shuffling would prevent beneficial mutations from being lost in false negatives, speeding directed evolution ever further.
The relationship between rounds of selection with semi-synthetic DNA shuffling, B2:B1 ratios in doramectin production, and number of mutations compared to wild-type aveC is compared in the following figure.
Conclusion
Semi-synthetic shuffling proved successful in quickly and efficiently improving the function of the aveC gene.
The Experiment
By conducting 5 rounds of semi-synthetic DNA shuffling on the aveC gene, the researchers were able to create a strain of S. avermitilis capable of producing doramectin with a B2:B1 ratio of 0.07:1.
To determine whether semi-synthetic DNA shuffling was effective at assembling multiple beneficial mutations in output strains, the most productive strains of each round of directed evolution were sequenced. While the best mutants from the first round had only five mutations, the most evolved mutations from the fifth round had X mutations. Thus there was a correlation between rounds of directed evolution and beneficial mutations in evolved aveC as well as rounds of directed evolution and decreases in the B2:B1 ratio.
Advantages of the Method
Semi-synthetic DNA shuffling seems to be a very effective method at speeding up the time required for satisfactory results from directed evolution.
Furthermore, semi-synthetic DNA shuffling allows rational input into directed evolution. If previous research has indicated that a certain mutation has a desirable effect on protein function, this mutation can be inserted into the DNA shuffling scheme as an oligonucleotide.
Disadvantages of the Method
The principal disadvantage of semi-synthetic DNA shuffling is that the method requires knowledge of gene to function relationship for the technique to work.
Its applicability to improvements of phenotypes regulated by many genes has not yet to be explored.
RETURN TO PAPER: Directed Evolution and Synthetic Biology - Hunter Stone