Shamita P
Cold Tolerance of the Northern highbush blueberry (Vaccinium corymbosum)
General background on cold tolerance
Winter Acclimation and Cold Hardiness of the Blueberry: Primarily geared towards individuals who wish to cultivate blueberries, but provides some good general background information on the cold tolerance associated with regional varieties.
Additionally, Polashock et al (2010) provides substantial background information on genetic basis of cold tolerance. In summary, they discuss that the purpose behind studying these genes is to understand how modifying cold-tolerance in blueberry might prevent massive crop loss due to freezing temperatures during a winter frost. The overall acclimation to cold occurs in two steps, the first of which is induced by a shorter photo-period (less sunlight), and the second of which is induced by lower temperatures. Polashock et al targeted a host of genes in a family of transcription factors called CBF (C-repeat binding factor). These TF appear to bind a conserved region CCGAC within promoters that activate a host of downstream genes involved in cold acclimation. Using this gene as a starting point, I decided to search for candidate genes downstream of CBFs in other species that were being activated in cold conditions.
Searching the CBF (C-repeat binding factor) genes
The exciting thing about CBFs is that they are found in many species of plants. So, if there are genes downstream of this TF in those plant species, they might be good targets for study in blueberry as well. I explored various papers discussing cold tolerance genes in Eucalyptus, Arabidopsis, and common wheat. Although common wheat is a monocot, I felt like it would be worth exploring because like blueberry, it is an important crop and might also have invested interest in its frost tolerance.
Starting with the paper above by Polashock et al, I obtained a list of the following genes from the following papers:
Cold Acclimation/Freezing Tolerance in Blueberries
Polashock et al (2010)
-COR6.6
-COR78
-COR15A etc..
Frost Tolerance in Temperate Cereals
Galiba et al (2009)
-FR2
-TaCBF14
-TaCBF15
Cold Tolerance in Eucalyptus Species
Navarro et al (2009)
-EguCBF1c
-EguCBF1d
Cold Tolerance signaling in Arabidopsis: ICE (Induction of CBF Expression)
Lissarre et al, 2010
-ICE1
-ICE2
CFB Genes
I decided to study at least one of the CBF transcription activators not found in blueberry as well as one frost tolerance gene found to be downstream of CBF. I searched the NCBI database to obtain mRNA sequences of my genes of interest. Of the several genes that I input into the Vaccinium database (conducting tBLASTx against the scaffolds), I found that there were two genes in particular with promising results. The first of these was the EguCBF1c in Eucalyptus, whose match against Scaffold 00009 had an E score of 10^(-31). When I submitted TaCBF14 gene from common wheat for analysis in the same manner, the top hit was also Scaffold 00009 with an E score of 10^(-17).
The tBLASTx translated my mRNA query into a protein and then matched it with all proteins constructed by all reading frames of the nucleotide sequence of the scaffolds. For this reasons, it's the longest search conducted in the BLAST database. Using the amino acid sequences that were output and their corresponding nucleotide matches to the scaffold, I was able to approximate where in the scaffold my genes were located. Both CBF genes Eucalyptus and Common wheat produced hits in the same region of the scaffold, at approximately 488,000 bp.
I submitted scaffold 00009 to be searched for SSRs using default parameters that favored lengthy di- or tri- nucleotide repeats. Vaccinium.org returns an excel file with the location and length of SSRs along with primers engineered to amplify the regions containing the SSR. See below.
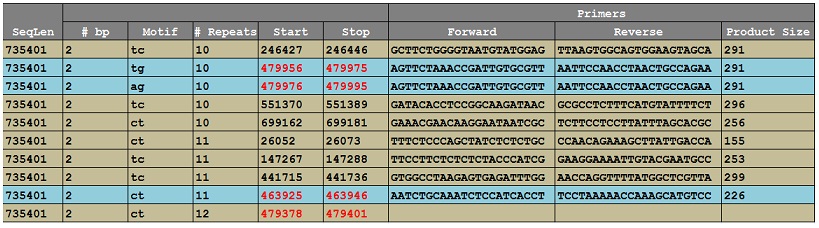
Choosing SSRs in the vicinity of my genes, I found 4 lengthy di-nucleotide repeats and one tri-nucleotide repeat around 488,000 bp (not pictured). The excel file does not always contain primers for every SSR match, so those positions are of no use to us. For primers it does provide, I chose ones that produced PCR products that were less than 300 bp.
When mapping this to the 282 pg Word File which contained the entire scaffold 00009, I found my SSR matches to be about 4 pages away from the 11 combined hits found from the gene search on the scaffold.
ICE1 Gene
Beginning with the tBLASTx search, I used the same steps with the ICE1 gene in Arabidopsis. The scaffold first hit on my search was Scaffold 00051 with an E score of 10^(-80). This is an extremely strong hit, that had 19 fragments of the ICE1 gene matching to the blueberry scaffold in high precision. All matches were between 55,000 and 60,000 bp on the scaffold (pg 22-23). I submitted Scaffold 00051 to the SSR database and found primers for two di-nucleotide and one tri-nucleotide repeats. Two of the primers were within the 5,000 bp range, while one was found at 67,000 bp.
SIZ1 Gene
Upon further reading, I found that ICE1 is activated by SIZ1 mediated SUMOylation. SUMOylation is a type of post-translational modification that involves the addition of a Small Ubiquitin-like Modifier (SUMO) to a protein, causing that protein to change its structure and thus its function. Among many reasons, SUMOylation is instigated in environments of stress such as freezing temperatures. I obtained the SIZ1 mRNA sequence for Arabidopsis and performed a tBLASTx on the sequence in the Vaccinium database. I found an exceptional match to scaffold 00717 (E = 0.0) and devised primers for this scaffold in the vicinity of the gene (85,000-107,000 bp). I found three good matches, whose PCR lengths and primers are shown below.
Possible Downstream Targets of CBFs
Results
EguCBF1c and TaCBF14
3 Primer Matches on Scaffold 00009 (~488,000 bp)
Forward Primer: AGTTCTAAACCGATTGTGCGTT
Reverse Primer: AATTCCAACCTAACTGCCAGAA
2-nucleotide repeat (20bp): 479,956 bp
Forward Primer: TCTCTCTCAGATCTCTGATCCGT
Reverse Primer: AAAGCAAGAAGAGAAATGGTGG
3-nucleotide repeat (15bp): 479,466 bp
Forward Primer: AATCTGCAAATCTCCATCACCT
Reverse Primer: TCCTAAAAACCAAAGCATGTCC
2-nucleotide repeat (22 bp): 463,925 bp
ICE1
3 Primer Matches on Scaffold 00051 (~55,000 - 60,000 bp)
Forward Primer: CGCATCTTTACTCCACTAACCC
Reverse Primer: AATCCCTGCTGTGTATCTTGGT
Di-nucleotide repeat (10bp): 55,088 bp
Forward Primer: GTGGGGAGCAAACTCACTAATC
Reverse Primer: AATAACAAAAACTCGCTCTCGC
Di-nucleotide repeat (10bp): 67,058 bp
Forward Primer: GAGAAGTGAAGGAATGGAGGTG
Reverse Primer: CGAAATGGGTTCACTCTCTACC
Tri-nucleotide repeat (12bp): 60,104 bp
SIZ1
3 Primer Matches on Scaffold 00717 (~85,000 - 107,000 bp)
Forward Primer: AAGCCGCATATTAGAGCGTATC
Reverse Primer: CCTCCCTCCTCTCTCTCTCTCT
Di-nucleotide repeat (42bp): 86,562 bp
Forward Primer: ATTGCAATCTTGCACAGAGAGA
Reverse Primer: CTACATAGGATACGCATTGGCA
Di-nucleotide repeat (10bp): 86,761 bp
Forward Primer: CATTTGTACCCCCTCAAGTAGC
Reverse Primer: TTTCCCTAGTGGTGAAGTGTGA
Tri-nucleotide repeat (12bp): 107,162 bp
IP5PII
3 Primer Matches on Scaffold 00661 (~93,000-105,000)
Forward Primer: GATTCGAACGGCAGTATAAACC
Reverse Primer: GCCCTTATCAATCTCCAAATGA
AT 6x @ 106,789, Product: 222 bp
Forward Primer: ATGGAGTACCAAGGAAAAACGA
Reverse Primer: CCATTTTTATCGGGGTGAGTAA
TC 13x @ 81,787, Product: 246 bp
Forward Primer: TCTCTTCTACTGTCAGAGGCCC
Reverse Primer: CACTCTGTTTGGAAAATGTGGA
ATA 5x @ 86,548, Product: 231 bp
ADTGK1
3 Primer Matches on Scaffold 00019 (~355,000-360,000)
Forward Primer: CTAGCCTACCAACTACCTCCGA
Reverse Primer: GGATTGCTTCTCTGTTTCTGCT
AG 7x @ 352,411, Product: 214 bp
Forward Primer: AGCAGAAACAGAGAAGCAATCC
Reverse Primer: CAAGGCAAACCCTAGAGAGAGA
CT 11x @ 352,582, Product: 143 bp
Forward Primer: TTGAACATGCTCTTGAATCCTG
Reverse Primer: TACGTGAGTATCATCCACAGCC
AATA 4x @ 355,495, Product: 131 bp