Difference between revisions of "Suppressor Logic Toolkit and Quasi-Hash Function"
m (→Basic Toolkit Design) |
(→Construct Design) |
||
| (38 intermediate revisions by 2 users not shown) | |||
| Line 14: | Line 14: | ||
== Toolkit with Specific Inputs and Outputs == | == Toolkit with Specific Inputs and Outputs == | ||
| − | PprpB/propionate and luxO/pluxT are new communication players that would need to be looked into further to verify their usefulness. | + | PprpB/propionate and luxO/pluxT or pqrr4 are new communication players that would need to be looked into further to verify their usefulness. |
See references at bottom of page. | See references at bottom of page. | ||
| Line 27: | Line 27: | ||
<center> [[Image:Picture21.gif]]<br></center> | <center> [[Image:Picture21.gif]]<br></center> | ||
<center> [[Image:Picture22.gif]]<br></center> | <center> [[Image:Picture22.gif]]<br></center> | ||
| + | |||
| + | == Construct Design == | ||
| + | |||
| + | '''NAND Gate construct''' | ||
| + | |||
| + | Plasmid 1 | ||
| + | |||
| + | pLuxR+RBS+5 bp suppressor+TT+pPrpB+RBS+modified cI+TT approx. 1552bp | ||
| + | |||
| + | Plasmid 2 | ||
| + | |||
| + | pLacI+RBS+LuxR+RBS+prpB+TT+pR+RBS+GFP+Mnt+TT+pMnt+RFP+TT approx. 4026bp | ||
| + | |||
| + | == 434 repressor == | ||
| + | ''Design:'' | ||
| + | |||
| + | 1. OR1 is needed. the role of OR1 is to help repressor bind to OR2 and to repress the PR; | ||
| + | |||
| + | 2. OR2 is needed. The binding of the '''amino-terminal domain''' of 434 repressor to OR2 is necessary and sufficient for activation of PRM transcription;[http://www.pnas.org/content/83/24/9353.full.pdf+html] | ||
| + | |||
| + | 3. Need to decrease the binding affinity of OR3. binding to OR3 will cause repression of PRM transcription.Introduce a mutation in OR3 that decreases its affinity for the repressor without disturbing PRM; | ||
| + | |||
| + | 4. Changing the sequence of the -35 region of PRM toward consensus greatly increases the ability of RNA polymerase to transcribe this DNA in the absence of the repressor...the mutation decreases the ability of RNA polymerase to initiate transcription at PR [http://www.jbc.org/cgi/content/full/273/37/24165] | ||
| + | |||
| + | http://gcat.davidson.edu/GcatWiki/images/c/ce/434r.JPG | ||
== Parts Needed == | == Parts Needed == | ||
| Line 36: | Line 61: | ||
pR [http://partsregistry.org/wiki/index.php?title=Part:BBa_R0051 R0051]2009 Kit Plate 1 6K pSB1A2 | pR [http://partsregistry.org/wiki/index.php?title=Part:BBa_R0051 R0051]2009 Kit Plate 1 6K pSB1A2 | ||
| − | pLacIQ1 [http://partsregistry.org/wiki/index.php/Part:BBa_K091112 K091112] | + | pLacIQ1 [http://partsregistry.org/wiki/index.php/Part:BBa_K091112 K091112]MWSU |
pTetR [http://partsregistry.org/wiki/index.php/Part:BBa_R0040 R0040]2009 Kit Plate 1 6I pSB1A2 | pTetR [http://partsregistry.org/wiki/index.php/Part:BBa_R0040 R0040]2009 Kit Plate 1 6I pSB1A2 | ||
| Line 56: | Line 81: | ||
RBS(B0030)+GFP [http://partsregistry.org/wiki/index.php/Part:BBa_E5500 E5500]2007 Parts Kit Plate 1 9J pSB1A2 | RBS(B0030)+GFP [http://partsregistry.org/wiki/index.php/Part:BBa_E5500 E5500]2007 Parts Kit Plate 1 9J pSB1A2 | ||
| − | RBS( | + | RBS(B0034)+RFP MWSU |
Mnt [http://partsregistry.org/wiki/index.php?title=Part:BBa_C0072 C0072]2009 Kit Plate 1 12L pSB2K3 | Mnt [http://partsregistry.org/wiki/index.php?title=Part:BBa_C0072 C0072]2009 Kit Plate 1 12L pSB2K3 | ||
LuxO-P [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131022 K131022]2009 Kit Plate 3 8K pSB1AC3 | LuxO-P [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131022 K131022]2009 Kit Plate 3 8K pSB1AC3 | ||
| + | |||
| + | pqrr4 [http://partsregistry.org/Part:BBa_K131017 K131017] | ||
| + | |||
| + | '''Parts that need modified''' | ||
| + | |||
| + | LuxO-P [http://partsregistry.org/wiki/index.php?title=Part:BBa_K131022 K131022]2009 Kit Plate 3 8K pSB1AC3 | ||
| + | |||
| + | RBS+lamba cI+TT [http://partsregistry.org/wiki/index.php/Part:BBa_P0451 P0451]2009 Kit Plate 2 1N pSB1AK3 | ||
'''Parts needed to be built from scratch''' | '''Parts needed to be built from scratch''' | ||
| − | + | '''prpBp''' [http://biocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3602 Ecocyc] | |
| − | + | (prpBp text version) | |
| − | + | tgaaattaaa catttaattt tattaaggca attgtggcac accccttgct ttgtctttat caacgcaaat aacaagttga taaca | |
| − | + | http://gcat.davidson.edu/GcatWiki/images/2/2f/PrpBp.JPG | |
| − | frame shift suppressor 2 | + | '''prpB''' 2-methylisocitrate lyase in E.coli MG1655 [http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=944990&ordinalpos=3&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum]http://gcat.davidson.edu/GcatWiki/images/6/6c/PrpB.JPG |
| + | |||
| + | (prpB text version) | ||
| + | |||
| + | atgTCTCTAC ACTCTCCAGG TAAAGCGTTT CGCGCTGCAC TGACTAAAGA AAATCCATTG | ||
| + | |||
| + | CAGATTGTTG GCACCATCAA CGCTAATCAT GCGCTGTTGG CGCAGCGTGC CGGATATCAG | ||
| + | |||
| + | GCAATTTATC TTTCTGGCGG TGGCGTGGCG GCAGGTTCGC TGGGGCTGCC CGATCTCGGT | ||
| + | |||
| + | ATTTCTACCC TTGATGATGT GCTGACCGAC ATTCGCCGTA TCACCGACGT TTGTTCGCTG | ||
| + | |||
| + | CCGCTGCTGG TGGATGCGGA TATCGGTTTT GGTTCTTCGG CCTTTAACGT GGCGCGCACC | ||
| + | |||
| + | GTGAAATCGA TGATTAAAGC CGGTGCGGCA GGATTGCATA TTGAAGATCA GGTTGGTGCG | ||
| + | |||
| + | AAACGCTGCG GTCATCGTCC GAATAAAGCG ATCGTCTCGA AAGAAGAGAT GGTGGATCGG | ||
| + | |||
| + | ATCCGCGCGG CGGTGGATGC GAAAACCGAT CCTGATTTTG TGATCATGGC GCGCACCGAT | ||
| + | |||
| + | GCTCTGGCGG TAGAGGGGCT GGATGCGGCG ATCGAGCGTG CGCAGGCCTA TGTTGAAGCG | ||
| + | |||
| + | GGTGCCGAGA TGTTGTTCCC GGAGGCGATT ACCGAACTCG CCATGTACCG CCAGTTTGCC | ||
| + | |||
| + | GATGCGGTGC AGGTGCCGAT CCTCGCCAAC ATCACCGAAT TTGGTGCCAC GCCGCTGTTT | ||
| + | |||
| + | ACCACCGACG AATTACGCAG CGCCCATGTC GCAATGGCGC TGTACCCACT TTCAGCGTTC | ||
| + | |||
| + | CGCGCCATGA ACCGCGCCGC TGAACATGTC TACAACGTCC TGCGCCAGGA AGGCACGCAG | ||
| + | |||
| + | AAAAGCGTCA TCGACACCAT GCAGACCCGC AACGAGCTGT ACGAAAGCAT CAACTACTAC | ||
| + | |||
| + | CAGTACGAAG AGAAGCTCGA CAACCTGTTT GCCCGTAGCC AGGTGAAATA A | ||
| + | |||
| + | '''pLuxT''' [http://www.jbc.org/cgi/reprint/281/46/34775] | ||
| + | http://gcat.davidson.edu/GcatWiki/images/e/e3/LuxT_upstream.JPG | ||
| + | |||
| + | (pLuxT text version) | ||
| + | |||
| + | agcgagcagt gagctcagtg gagtcatcag ttaaatctgt agtgttgtat tccacatttt | ||
| + | |||
| + | cgtttactgc tttcgccagc tctagatact tggcaaattc cgcttgtagc aattcttttg | ||
| + | |||
| + | aaccagcgta aatcgtcacg tctgcaacgt catcgccttc tttgatgata aagccaattt | ||
| + | |||
| + | caccagcgca accacacgct tcacatactt gggtttcttc gtgttcaatg ctcataacga | ||
| + | |||
| + | atcctcactt aaaaaaggga ttatactcaa ctcgtttttt tttcaataga aataaatgga | ||
| + | |||
| + | acaacttgga taaaaaaata tcggtatgct atgttgtttg ttgagagggt taagcaagag | ||
| + | |||
| + | ataactttag attttgttaa tggacgatgt gatccgagtc aataatttac cggatttcac | ||
| + | |||
| + | cttttactta cttagttatc acagtgagtg agttgcacct agcaagattt tataagagta | ||
| + | |||
| + | tctcgagata aattattttt acataatcta tcgctatcaa tcggttcaaa atgcagatgt | ||
| + | |||
| + | aaatattaaa ctaatgtcat ttaatggtat taatttagtt gcatattctg aaattttgtt | ||
| + | |||
| + | ttgcttgaag acatgcatct gcaaacctgg tttacagtat ttcgtaataa aaagtaggct | ||
| + | |||
| + | tgtttatttc gaaaaagggt gttttctggc aacctattga gaacgatttt cattctgacc | ||
| + | |||
| + | tcagactttg agttagatgg atttacaaag agaagagcta aaatatgcca aagcgtagta | ||
| + | |||
| + | aagaagatac cgaaatcact atccagaaaa ttatggatgc cgttgttgat cagttgctta | ||
| + | |||
| + | ggctcggtta cgacaaaatg tcgtacacga cgttgagtca acagacaggt gtttcacgta | ||
| + | |||
| + | (Xba1 site at bp81) | ||
| + | |||
| + | '''frame shift suppressor 1''' CU'''GUCCU'''AA | ||
| + | |||
| + | '''frame shift suppressor 2''' UU'''ACUAG'''AC | ||
| + | |||
| + | prpBp and prpB have no EcoR1, Xba1, Spe1, or Pst1 sites. Source www.neb.com | ||
== References == | == References == | ||
| Line 83: | Line 191: | ||
PprpB propionate [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16269719] | PprpB propionate [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=16269719] | ||
| + | |||
| + | 434 repressor [http://www.pnas.org/content/83/24/9353.full.pdf+html] [http://www.jbc.org/cgi/content/full/273/37/24165] | ||
Latest revision as of 14:09, 29 July 2009
Contents
Suppressor Logic Toolkit and Quasi-Hash Function
The following slides describe the basic toolkit design and a specific experiment that would result in a quasi-hash function
Basic Toolkit Design
The AND gate was designed by Anderson using termination suppressors, while the new design would use frame shift suppressors.
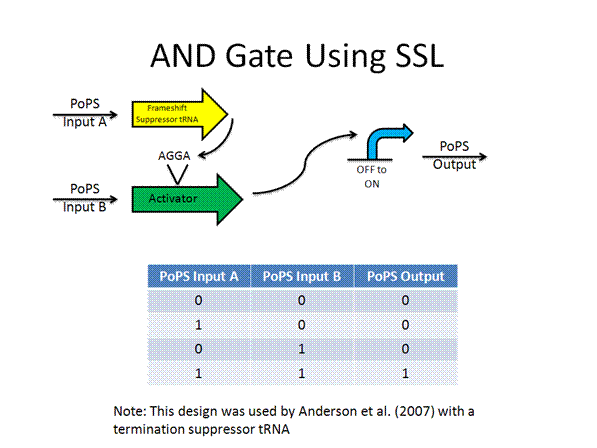
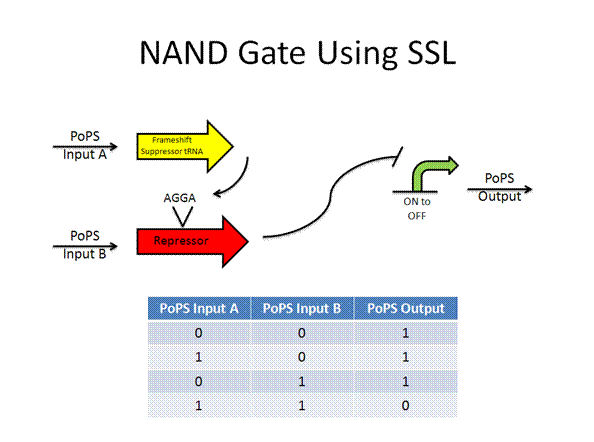
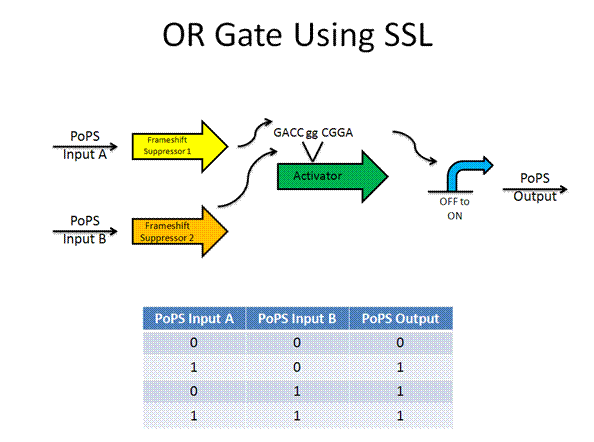
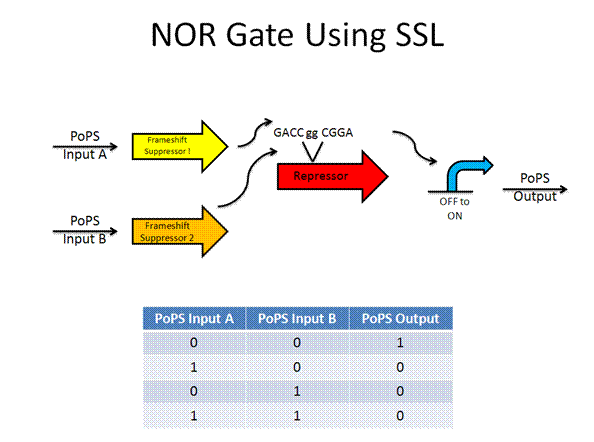
Toolkit with Specific Inputs and Outputs
PprpB/propionate and luxO/pluxT or pqrr4 are new communication players that would need to be looked into further to verify their usefulness.
See references at bottom of page.
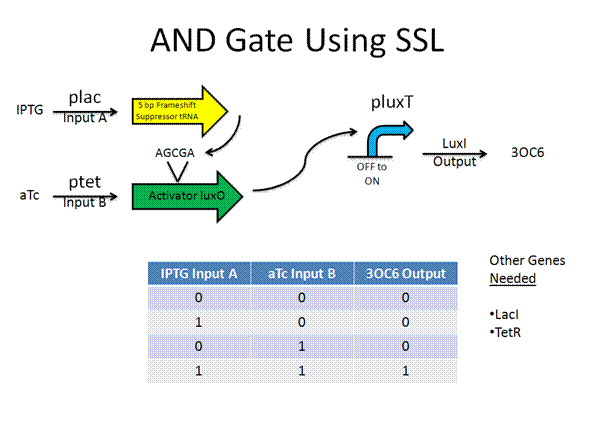
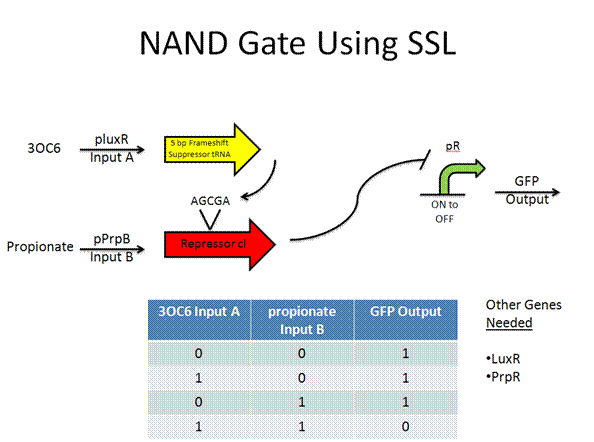
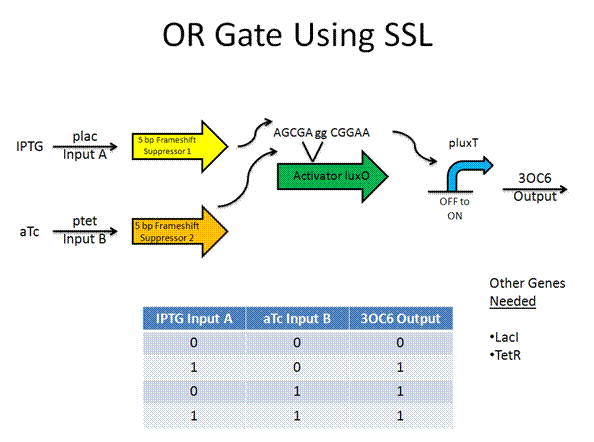
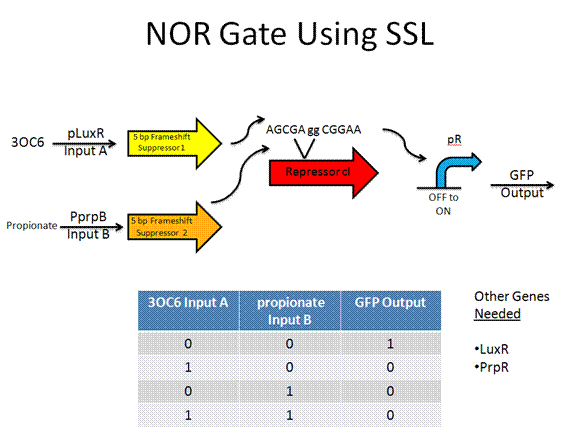
Experimental Design
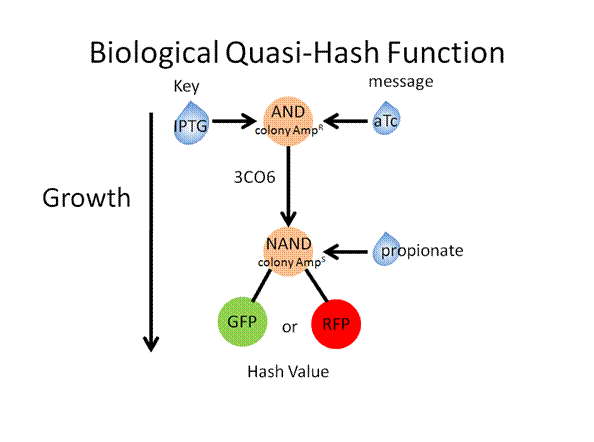
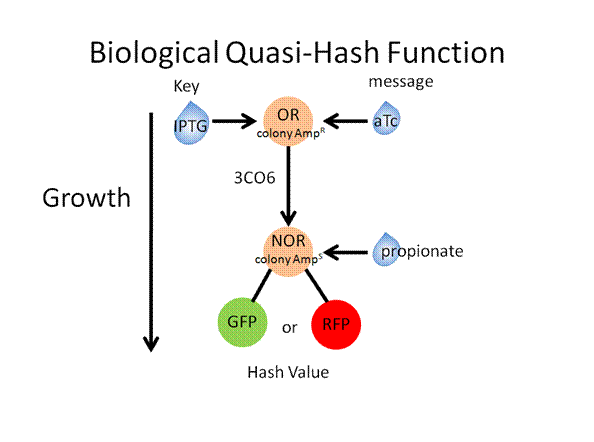
Construct Design
NAND Gate construct
Plasmid 1
pLuxR+RBS+5 bp suppressor+TT+pPrpB+RBS+modified cI+TT approx. 1552bp
Plasmid 2
pLacI+RBS+LuxR+RBS+prpB+TT+pR+RBS+GFP+Mnt+TT+pMnt+RFP+TT approx. 4026bp
434 repressor
Design:
1. OR1 is needed. the role of OR1 is to help repressor bind to OR2 and to repress the PR;
2. OR2 is needed. The binding of the amino-terminal domain of 434 repressor to OR2 is necessary and sufficient for activation of PRM transcription;[1]
3. Need to decrease the binding affinity of OR3. binding to OR3 will cause repression of PRM transcription.Introduce a mutation in OR3 that decreases its affinity for the repressor without disturbing PRM;
4. Changing the sequence of the -35 region of PRM toward consensus greatly increases the ability of RNA polymerase to transcribe this DNA in the absence of the repressor...the mutation decreases the ability of RNA polymerase to initiate transcription at PR [2]
http://gcat.davidson.edu/GcatWiki/images/c/ce/434r.JPG
Parts Needed
Parts from Registry
Promoters
pR R00512009 Kit Plate 1 6K pSB1A2
pLacIQ1 K091112MWSU
pTetR R00402009 Kit Plate 1 6I pSB1A2
pLuxR R00622009 Kit Plate 1 6O pSB1A2
pMnt R00732009 Kit Plate 1 16B pSB2K3
Genes
RBS+lamba cI+TT P04512009 Kit Plate 2 1N pSB1AK3
pBAD+RBS+LacI+TT S03980MWSU
pBAD+RBS+tetR+TT K091145MWSU
pBAD+RBS+luxR+TT K091179MWSU
RBS(B0030)+GFP E55002007 Parts Kit Plate 1 9J pSB1A2
RBS(B0034)+RFP MWSU
Mnt C00722009 Kit Plate 1 12L pSB2K3
LuxO-P K1310222009 Kit Plate 3 8K pSB1AC3
pqrr4 K131017
Parts that need modified
LuxO-P K1310222009 Kit Plate 3 8K pSB1AC3
RBS+lamba cI+TT P04512009 Kit Plate 2 1N pSB1AK3
Parts needed to be built from scratch
prpBp Ecocyc
(prpBp text version)
tgaaattaaa catttaattt tattaaggca attgtggcac accccttgct ttgtctttat caacgcaaat aacaagttga taaca
http://gcat.davidson.edu/GcatWiki/images/2/2f/PrpBp.JPG
prpB 2-methylisocitrate lyase in E.coli MG1655 [3]http://gcat.davidson.edu/GcatWiki/images/6/6c/PrpB.JPG
(prpB text version)
atgTCTCTAC ACTCTCCAGG TAAAGCGTTT CGCGCTGCAC TGACTAAAGA AAATCCATTG
CAGATTGTTG GCACCATCAA CGCTAATCAT GCGCTGTTGG CGCAGCGTGC CGGATATCAG
GCAATTTATC TTTCTGGCGG TGGCGTGGCG GCAGGTTCGC TGGGGCTGCC CGATCTCGGT
ATTTCTACCC TTGATGATGT GCTGACCGAC ATTCGCCGTA TCACCGACGT TTGTTCGCTG
CCGCTGCTGG TGGATGCGGA TATCGGTTTT GGTTCTTCGG CCTTTAACGT GGCGCGCACC
GTGAAATCGA TGATTAAAGC CGGTGCGGCA GGATTGCATA TTGAAGATCA GGTTGGTGCG
AAACGCTGCG GTCATCGTCC GAATAAAGCG ATCGTCTCGA AAGAAGAGAT GGTGGATCGG
ATCCGCGCGG CGGTGGATGC GAAAACCGAT CCTGATTTTG TGATCATGGC GCGCACCGAT
GCTCTGGCGG TAGAGGGGCT GGATGCGGCG ATCGAGCGTG CGCAGGCCTA TGTTGAAGCG
GGTGCCGAGA TGTTGTTCCC GGAGGCGATT ACCGAACTCG CCATGTACCG CCAGTTTGCC
GATGCGGTGC AGGTGCCGAT CCTCGCCAAC ATCACCGAAT TTGGTGCCAC GCCGCTGTTT
ACCACCGACG AATTACGCAG CGCCCATGTC GCAATGGCGC TGTACCCACT TTCAGCGTTC
CGCGCCATGA ACCGCGCCGC TGAACATGTC TACAACGTCC TGCGCCAGGA AGGCACGCAG
AAAAGCGTCA TCGACACCAT GCAGACCCGC AACGAGCTGT ACGAAAGCAT CAACTACTAC
CAGTACGAAG AGAAGCTCGA CAACCTGTTT GCCCGTAGCC AGGTGAAATA A
pLuxT [4] http://gcat.davidson.edu/GcatWiki/images/e/e3/LuxT_upstream.JPG
(pLuxT text version)
agcgagcagt gagctcagtg gagtcatcag ttaaatctgt agtgttgtat tccacatttt
cgtttactgc tttcgccagc tctagatact tggcaaattc cgcttgtagc aattcttttg
aaccagcgta aatcgtcacg tctgcaacgt catcgccttc tttgatgata aagccaattt
caccagcgca accacacgct tcacatactt gggtttcttc gtgttcaatg ctcataacga
atcctcactt aaaaaaggga ttatactcaa ctcgtttttt tttcaataga aataaatgga
acaacttgga taaaaaaata tcggtatgct atgttgtttg ttgagagggt taagcaagag
ataactttag attttgttaa tggacgatgt gatccgagtc aataatttac cggatttcac
cttttactta cttagttatc acagtgagtg agttgcacct agcaagattt tataagagta
tctcgagata aattattttt acataatcta tcgctatcaa tcggttcaaa atgcagatgt
aaatattaaa ctaatgtcat ttaatggtat taatttagtt gcatattctg aaattttgtt
ttgcttgaag acatgcatct gcaaacctgg tttacagtat ttcgtaataa aaagtaggct
tgtttatttc gaaaaagggt gttttctggc aacctattga gaacgatttt cattctgacc
tcagactttg agttagatgg atttacaaag agaagagcta aaatatgcca aagcgtagta
aagaagatac cgaaatcact atccagaaaa ttatggatgc cgttgttgat cagttgctta
ggctcggtta cgacaaaatg tcgtacacga cgttgagtca acagacaggt gtttcacgta
(Xba1 site at bp81)
frame shift suppressor 1 CUGUCCUAA
frame shift suppressor 2 UUACUAGAC
prpBp and prpB have no EcoR1, Xba1, Spe1, or Pst1 sites. Source www.neb.com
References
Powerpoint File:Suppressorlogic3.ppt
Anderson et al. 2007. “Environmental signal integration by a modular AND gate” [5]
luxO luxT [6]
PprpB propionate [7]