Difference between revisions of "TBLASTn and Protein Sequence Analysis"
(→When Nucleotides Don't Work) |
Macampbell (talk | contribs) |
||
| (22 intermediate revisions by one other user not shown) | |||
| Line 10: | Line 10: | ||
But assuming that you're working with a gene that should be highly conserved and you have allowed for the exon/intron issue, it may be time to go through some extra steps to make sure everything is working as it should: | But assuming that you're working with a gene that should be highly conserved and you have allowed for the exon/intron issue, it may be time to go through some extra steps to make sure everything is working as it should: | ||
| − | + | :'''''First''''' -- Check to make sure that your database is made correctly. | |
| − | + | ::*You should have three additional files made from the original fasta file: -.nsq, -.nin, and -.nhr | |
| − | + | :'''''Second''''' -- Check that your commands are entered correctly. | |
| − | + | ::*Ensure that you are in the correct directory (i.e. Desktop) and BLASTing with the correct file name | |
| − | + | ::*Sequences being blasted should be in TextEdit and saved without any extension (such as .txt) | |
| − | + | :'''''Third''''' -- Ensure that the databases/commands are working. | |
| − | + | ::*Take a known sequence from the database being blasted | |
| − | + | :::*Typically copy a small portion of the genome into a separate text file | |
| − | + | ::*Run a BLAST with that sequence against the database it was originally in | |
| − | + | :::*If the sequence doesn't show a hit in the scaffold that you took it from, something is wrong in your programming | |
If you've gone through these steps and still can't find the issue, it might be time to move away from nucleotide sequences and try some amino acid sequences. | If you've gone through these steps and still can't find the issue, it might be time to move away from nucleotide sequences and try some amino acid sequences. | ||
| − | == | + | == Protein BLASTing == |
| − | == Why | + | === Why BLAST proteins? === |
| + | Working at the protein level provides a number of benefits. Because each amino acid can be coded for by 3 or 4 different codons, a given gene could have a number of different nucleotide sequences that all produce the same amino acids. Therefore, BLASTing at the protein level allows for a certain flexibility that cannot be achieved with nucleotides alone. Protein BLASTing will also account for any ''silent mutations'' (point mutations that do not change amino acid sequence) present in the genome. | ||
| + | |||
| + | === Conserved Protein Sequences === | ||
| + | Even in highly conserved proteins, there will be some variation across species and individuals. Oftentimes, however, there is at least one region of the gene that had an important enough function that it changed very little over time. These especially conserved portions of the protein can be very useful when trying to identify them in a new genome. | ||
| + | |||
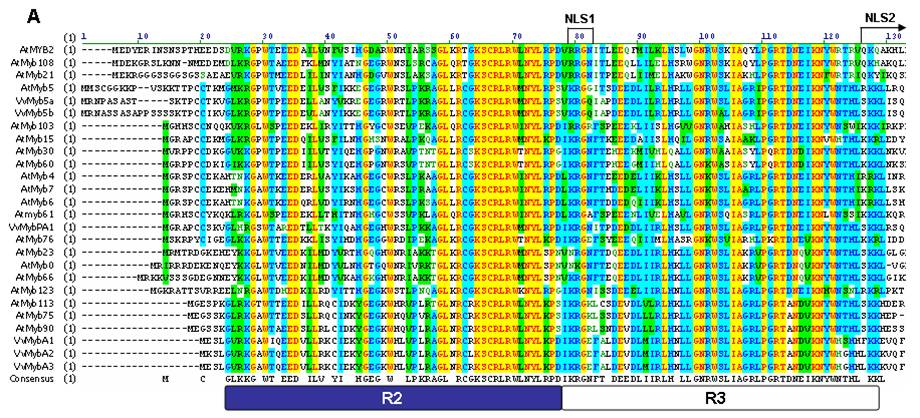
| + | The Myb transcription factor family in plants provides a perfect example of a protein that has one highly conserved region. This is known as the Myb Domain and determines how the protein binds to DNA. When a number of these proteins are compared with one another, it is possible to see where these regions exist. The graphic below was created by a group looking for Myb sequences in the grape genome. | ||
| + | |||
| + | [[File:Conserved Myb.jpg]][http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2507771/?tool=pubmed#S1] | ||
| + | |||
| + | This table shows a series of known Myb proteins from the ''Arabidopsis'' genome. By doing so, they were able to parse out which parts of the protein sequence are the highly conserved. This can be seen on the last line, labeled "Consensus". | ||
| + | |||
| + | === Using tBLASTn === | ||
| + | Once a protein consensus sequence has been obtained, it can be BLASTed against the genome in question. This is done through the use of a tBLASTn. Ordinary queries use a BLASTn. This means that the program searches for a nucleotide sequence within a nucleotide database. There are numerous programs that can be used to BLAST depending on the intent and nature of the search. Descriptions of all such programs can be found at the [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=ProgSelectionGuide NCBI Blast Program Selection Guide]. | ||
| + | |||
| + | |||
| + | tBLASTn works in much the same way that BLASTn does, but adds in an extra step. Before BLASTing, the tBLASTn program must convert the database of nucleotide sequences into a amino acid sequences. tBLASTn will query with your protein sequence against the translated nucleotide sequences. tBLASTn allows any genes with silent mutations to be detected as an ortholog. | ||
| + | |||
| + | |||
| + | In order to use the tBLASTn program, only a few basic changes are necessary to the standard BLAST program: | ||
| + | :*Ensure that the TextEdit file contains an amino acid sequence. Remove any "Met" designations and replace with "M". Also be sure that there are no stop codons in the sequence. | ||
| + | :*Replace the '''blastn''' in the command with '''tblastn''' | ||
| + | ::Normal BLAST ''/usr/local/ncbi/blast/bin/'''blastn''' -query FILE_NAME -db DATABASE_NAME -outfmt "7 qacc sacc evalue qstart qend sstart send"'' | ||
| + | ::Protein BLAST ''/usr/local/ncbi/blast/bin/'''tblastn''' -query FILE_NAME -db DATABASE_NAME -outfmt "7 qacc sacc evalue qstart qend sstart send"'' | ||
| + | |||
| + | === Interpreting Your Results === | ||
| + | Results are provided in exactly the same way that an ordinary BLAST would output. The location and e-value of each hit is provided. This makes it easy to establish the validity of the find and actually locate the gene sequence within the genome. | ||
| + | |||
| + | |||
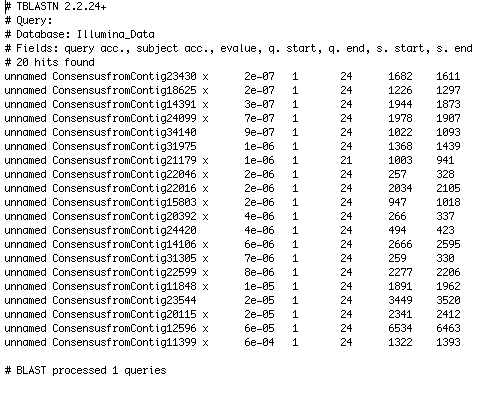
| + | The example below provides the result of when the Myb Consensus Sequence shown above was BLASTed against a new genome. | ||
| + | |||
| + | [[File:Screenshot.jpg]] | ||
| + | |||
| + | As can be seen, the Contig and nucleotide range are provided for easy location and the e-value shows how strong the hit is. | ||
Latest revision as of 14:53, 13 February 2013
Contents
When Nucleotides Don't Work
Occasionally BLASTing genes with just a nucleotide sequence doesn't work. This can be due to a number of reasons:
- Lack of conservation between species
- Incomplete genome database
- Rearrangement of introns/exons
But assuming that you're working with a gene that should be highly conserved and you have allowed for the exon/intron issue, it may be time to go through some extra steps to make sure everything is working as it should:
- First -- Check to make sure that your database is made correctly.
- You should have three additional files made from the original fasta file: -.nsq, -.nin, and -.nhr
- Second -- Check that your commands are entered correctly.
- Ensure that you are in the correct directory (i.e. Desktop) and BLASTing with the correct file name
- Sequences being blasted should be in TextEdit and saved without any extension (such as .txt)
- Third -- Ensure that the databases/commands are working.
- Take a known sequence from the database being blasted
- Typically copy a small portion of the genome into a separate text file
- Run a BLAST with that sequence against the database it was originally in
- If the sequence doesn't show a hit in the scaffold that you took it from, something is wrong in your programming
If you've gone through these steps and still can't find the issue, it might be time to move away from nucleotide sequences and try some amino acid sequences.
Protein BLASTing
Why BLAST proteins?
Working at the protein level provides a number of benefits. Because each amino acid can be coded for by 3 or 4 different codons, a given gene could have a number of different nucleotide sequences that all produce the same amino acids. Therefore, BLASTing at the protein level allows for a certain flexibility that cannot be achieved with nucleotides alone. Protein BLASTing will also account for any silent mutations (point mutations that do not change amino acid sequence) present in the genome.
Conserved Protein Sequences
Even in highly conserved proteins, there will be some variation across species and individuals. Oftentimes, however, there is at least one region of the gene that had an important enough function that it changed very little over time. These especially conserved portions of the protein can be very useful when trying to identify them in a new genome.
The Myb transcription factor family in plants provides a perfect example of a protein that has one highly conserved region. This is known as the Myb Domain and determines how the protein binds to DNA. When a number of these proteins are compared with one another, it is possible to see where these regions exist. The graphic below was created by a group looking for Myb sequences in the grape genome.
This table shows a series of known Myb proteins from the Arabidopsis genome. By doing so, they were able to parse out which parts of the protein sequence are the highly conserved. This can be seen on the last line, labeled "Consensus".
Using tBLASTn
Once a protein consensus sequence has been obtained, it can be BLASTed against the genome in question. This is done through the use of a tBLASTn. Ordinary queries use a BLASTn. This means that the program searches for a nucleotide sequence within a nucleotide database. There are numerous programs that can be used to BLAST depending on the intent and nature of the search. Descriptions of all such programs can be found at the NCBI Blast Program Selection Guide.
tBLASTn works in much the same way that BLASTn does, but adds in an extra step. Before BLASTing, the tBLASTn program must convert the database of nucleotide sequences into a amino acid sequences. tBLASTn will query with your protein sequence against the translated nucleotide sequences. tBLASTn allows any genes with silent mutations to be detected as an ortholog.
In order to use the tBLASTn program, only a few basic changes are necessary to the standard BLAST program:
- Ensure that the TextEdit file contains an amino acid sequence. Remove any "Met" designations and replace with "M". Also be sure that there are no stop codons in the sequence.
- Replace the blastn in the command with tblastn
- Normal BLAST /usr/local/ncbi/blast/bin/blastn -query FILE_NAME -db DATABASE_NAME -outfmt "7 qacc sacc evalue qstart qend sstart send"
- Protein BLAST /usr/local/ncbi/blast/bin/tblastn -query FILE_NAME -db DATABASE_NAME -outfmt "7 qacc sacc evalue qstart qend sstart send"
Interpreting Your Results
Results are provided in exactly the same way that an ordinary BLAST would output. The location and e-value of each hit is provided. This makes it easy to establish the validity of the find and actually locate the gene sequence within the genome.
The example below provides the result of when the Myb Consensus Sequence shown above was BLASTed against a new genome.
As can be seen, the Contig and nucleotide range are provided for easy location and the e-value shows how strong the hit is.