Difference between revisions of "Summer 2012 SynBio Project (Davidson and MWSU)"
(→GGA for College Teaching Labs) |
(→GGA for College Teaching Labs) |
||
| Line 498: | Line 498: | ||
[[File:1µL10cycles.jpg|400px|]]<br> | [[File:1µL10cycles.jpg|400px|]]<br> | ||
'''Figure 5:''' Photograph of 1 µL unclean PCR product plate, 10 cycles | '''Figure 5:''' Photograph of 1 µL unclean PCR product plate, 10 cycles | ||
| + | <br> | ||
| + | [[File:5µL5cycles.jpeg|400px|]]<br> | ||
| + | '''Figure 6:''' Photograph of 5 µL unclean PCR product plate, 5 cycles | ||
| + | <br> | ||
Revision as of 14:15, 2 July 2012
- Davidson Protocols
- MWSU_protocols
- GCAT-alog Freezer Stocks
- Laboratory_Notebooks
- Golden Gate
- Philosophy and Ethics of our Project
Contents
Student Proposals from Ind. Studies
- Erich Baker Proposal: Media:Erich_Baker_proposal.docx This proposal deals with Phytochromes and Light Sensitive Channel Proteins
-I think the use of Phytochromes might be a good way to have either a continual stimulus that would repress/express certain genes that could be turned off and on depending on what we want them to do. There are other aspects of the research in this proposal that if not used outright, could be adapted to our continuing projects as either controls or feedback mechanisms. As for the proposed Salis RBS sites, I would like to see more information in the efficacy of the predicted RBS sequence. Possibly if we could use some of the C-Dog information based on a few known sequences to determine if the computer can predict those RBS's we know to be effective then we might be able to count on the calculator as a tool for our experimental design. -Caleb Carr
- Ben Clarkson Proposal: Media:Ben_Clarkson_proposal.docx
- Duke DeLoache Proposal: Media:Duke_DeLoache_Proposal.docx
- Becca Evans Proposal: Media:Becca_Evans_proposal.docx
- Ellen Johnson Proposal: Media:Ellen_Johnson_proposal.docx
PPT Presentations
- This PPT file contains all the slides from student presentations addressing the idea proposed by MWSU.
Media:Reports_on_Circuits.pptx
- This PPT contains slides summarizing some of the best and most complicated papers from Week 11.
Papers
Methods Papers
- DNA assembly for synthetic biology: from parts to pathways and beyond
Tom Ellis, Tom Adieac and Geoff S. Baldwin
Integr. Biol., 2011, 3, 109–118
- Everyone should watch this 5 minute video on optogenetics. Combine that video with the 2010 champoinship iGEM invention of E. glowi.
Older Lab Papers
- Engineering bacteria to solve the Burnt Pancake Problem.
Haynes, Karmella, et al.
Journal of Biological Engineering. Vol. 2(8): 1 – 12.
- Solving a Hamiltonian Path Problem with a Bacterial Computer.
Baumgardner, Jordan et al.
Journal of Biological Engineering. Vol. 3:11
- Bacterial Hash Function Using DNA-Based XOR Logic Reveals Unexpected Behavior of the LuxR Promoter.
Brianna Pearson*, Kin H. Lau* et al.
Interdisciplinary Bio Central. Vol. 3, article no. 10.
Time Delayed Growth Movie
Network Papers
Jonathan M. Raser and Erin K. O’Shea
Science. Vol. 309, page 2010
Please post pdf.
Nagarajan Nandagopal and Michael B. Elowitz
Science. Vol. 333, page 1244.
Please post pdf.
R. Milo, S. Shen-Orr, et al
Science. Vol. 298, page 824.
Please post pdf.
Yang-Yu Liu, Jean-Jacques Slotine, & Albert-La ́szlo ́ Baraba ́si
Nature. 2011. Vol. 473, page 167.
Please post pdf.
Ethics Papers
Colin Mcilswain
Nature. Vol 465, page 867.
- Word selection affects perceptions of synthetic biology. Brianna Pearson, Sam Snell, Kyri Bye-Nagel, Scott Tonidandel, Laurie J Heyer, and A Malcolm Campbell.
-This paper does a great job at highlighting the importance of socio-political legitimation in the funding of science. It seems that all new sciences must survive a period during which their only funding comes from public sources under the condition that those conducting it can make some kind of promises of future benefit to the society as a whole. After proving itself not only useful but also profitable, private money may then start flowing in, though by that point, the nature of that field may arguably have changed for better or worse. I think we would all agree that synthetic biology holds more promise than we can currently even imagine, both for advancing the public good and for providing opportunity for profit (in more than just pharmaceuticals), but it's not enough for us to believe it. Those of us who will someday pursue grants and/or private investments in synthetic biology must learn to speak not only the rational language of the science of synthetic biology but also the politically-driven language of the social benefits of synthetic biology, the socially conscious language of the ethics of synthetic biology, and the profit-driven language of the (future) business of synthetic biology (and possibly others). -Eddie Miles
- Read "Moral" ethics paper on synthetic biology. Media:Moral.pdf
- Read "Future" ethics paper on synthetic biology. Media:Future.pdf
Questions to Consider About Network Pathways
- Are they naturally occurring or synthetic?
- Do they involve screening or selection?
- Are they anabolic or catabolic?
- How many steps are in each pathway?
- How can they relate to cell fitness?
- What specific challenges would need to be addressed if we worked with the pathway?
Cellular Automata
- [1] General CA introduction
- [2], [3] Elementary Cellular Automata
- [4] Good explanation of how elementary CAs work
- [5] Rule 110
- iGEM Team Groningen
- iGEM Team IPN-UNAM Mexico
- MIT 2011 iGEM Tissue Design
- In Vivo Cellular Automata
- Edge Detection PDF
- Patterning of E. coli
- Tunable Bacterial Band-Pass Filter
- E. coli Predator-Prey Ecosystem
- Multicellular System for Programmed Pattern Formation
Peptides
- Pep-1 can carry large amounts of cargo across cell membrane
- Pep-1 has no anti-microbial activity against E. coli, see page 121
- [6]General Manual for CPP After opening, click on the PDF General Manual for detailed information concerning Cellular Permeating Peptides, and products of the like.
- General info on Pep-1
- Very clear, easy to read, discussion on how CPPs work, and more specific info on Pep-1, look in Chapter 1 to start
- Pep-1 is a synthetic peptide
- Pep-1 fusion protein made in E. coli
- Targeting proteins to E. coli periplasmic space (GFP)
- Review of targeting proteins to periplasm
Environmental factors that enhance the action of the cell penetrating peptide pep-1 - A spectroscopic study using lipidic vesicles [[7]]
Assembly
[8]iGEM Introduction to Gibson Assembly
[9]Enzymatic assembly of DNA molecules up to several hundred kilobases
[10] Supplemental Methods for Enzymatic assembly of DNA molecules up to several hundred kilobases
[11]Assembly of BioBricks by the Gibson Method
[12] Properties of Exonuclease
[13]Tool for using Gibson Assembly Method
Library of Parts
Research Papers, Articles & Manuscripts--all inclusive and in regards to any and all parts that are listed, or wish to be listed
- [14] gene-specific promoter element is required for optimal expression of the histone H1 gene in S-phase.
- [15] Multiple Sigma Factors
Promoters Section
- 6 possible promoters for project 3 constitutive, 3 inducible - (Word file not yet saved on wiki)
C-Dog Section
Degradation Tag Section
Selection Modules
Bad-ish genes/proteins
- Toxicity of rat insulin gene on E.coli
- SacB gene with sucrose and E.coli
- Hda-mediated homeostasis in E.coli
- Lon protease from M.smegmatis
- SinI enzyme has moderate growth-inhibition in E.coli
- Excess violecein production toxic to E.coli
- ToxN inhibits growth of E.coli
- Eukaryotic membrane proteins toxic to E.coli
- HbpA proteins moderately toxic to E.coli
- Alpha-luffin and E.coli
- BBG29 gene from 'Borrelia'
Good genes/proteins
CRISPR process
- Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli
- Envelope stress trigger for CRISPR response - and background info on E.coli Cascade complex
- CRISPR database - to compare and find
- CRISPR immune system in Sulfolobales
- CRISPR System protects Microbes against Phages, Plasmids
- Impact on Small Repeat Sequences on Bacterial Genome Evolution
- Structural Basis for CRISPR RNA-guided DNA recognition by Cascade
- Structures of the RNA-guided surveillance complex from a bacterial immune system -figures of Subnanometer structures of Cascade
- 2011 iGEM team- CRISPR/Cas and GFP
- Diversity of CRISPR loci in E.coli
- Guild of 45 CRISPR-associated protein families
- CRISPR Interference Directs Strand Specific Spacer Acquisition
- CRISPR interference: RNA-directed Adaptive Immunity in Bacteria and Archaea
- Mature crRNA length measured by ruler mechanism
- CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation
- Media:CRISPR-based adaptive immune systems-terns.pdf
- Media:Evolution and Classification of the CRISPR-Cas systems- Makarova.pdf
- Media:Essential features and rational design of CRISPR RNAs that function with the Cas RAMP Module complex to cleave RNAs.pdf
- Media:Supplemental information to Essential Features paper.pdf
- The CRISPR System: small RNA-guided defense in bacteria and archaea
- The Escherichia coli CRISPR System Protects from lambda Lysogenization, Lysogens, and Prophage Induction
- Short motif sequences determine the targets of the prokaryotic CRISPR defense system
- The Small, Slow, and Specialized CRISPR and Anti-CRISPR of Escherichia and Salmonella
Regulated Biosynthesis Pathways
http://cat.inist.fr/?aModele=afficheN&cpsidt=6828850
Aptamers
- Use of riboswitch in Bacteria
- Screen for synthetic riboswitches reveals mechanistic insights into their function
- Riboswitches: A Common RNA Regulatory Element
Gas-Phase Communication
- Biopixel Paper
- ArcAB system in V. fischeri Includes promoter sequences
- ArcAB system in E. coli
- ArcAB system Responses to Hydrogen Peroxide in E. coli
- Visual Diagram of ArcAB System in E. coli
- Amino Acid Sequence for ArcA in E. coli
- Amino Acid Sequence for ArcB in E. coli
- Amino Acid Sequence for ArcA in V. Fischeri
- Blast Amino Acid Sequence Comparison for ArcA in E. coli and V. fischeri
- Blast Amino Acid Sequence Comparison for ArcB in E. coli and V. fischeri
- Potential Promoters that ArcA Might Bind To in E. coli
- More Potential Promoters that ArcA Might Bind To in E. coli-fad regulon
- List of Operons Repressed or Activated by ArcA in E. coli
- Sequence of focAP2 promoter-activated by ArcA
- Sequence of cydAP1 promoter-activated by ArcA
- Sequence of icdAp1 promoter-repressed by ArcA
- CydAB Activated in E. coli
- Sub-lethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis
- ArcAB system and how it works-sort of
- Specifics of how ArcB works/its composition
- How ArcAB functions as resistance to reactive oxygen stress/hydrogen peroxide
- ArcAB and the cydAB promoter with the H-NS protein
- The Effects of D-lactate on ArcB in Aerobic and Anaerobic Conditions
- Intracellular Concentration of Hydrogen Peroxide and Catalase in E. coli
Light
- light-gated ion channels/pumps
| Pump | phR | MscL | NpHR | e-BO/e-BR/h-BR | PR |
|---|---|---|---|---|---|
| wavelength | max absorbance at 578-599 | open with 366 nm, close with visible light (>466 nm) | 578 nm (with NaCl in media) | 550-560 nm | ~525 nm |
| particles that can travel through it | Chloride ions | non-selective, 3-nm diameter | anions | protons | protons |
| pump/channel? | pump | channel | pump | pump | pump |
| type of protein | halorhodopsin | n/a | halorhodopsin | BR=bacteriorhodopsin, BO=bacterio-opsin | proteorhodopsin |
| direction | into cell | n/a | into cell | into cell | out of cell |
- pH inducible promoters
- pH inducible promoter system pSM10
- Media:pH inducible promoter.pdf pSM-10
- High pH induced proteins
- more high/low pH induced proteins/genes
- high/low pH induced proteins/genes (more)
- TnaA
- cpxP (high ph induced)
- cadA promoter sequence (low pH induced)
- rest of cadA promoter sequence
- Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia ecoli cadBA operon
- Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH.
- File:Küper and Jung 2005.pdf Cad1 and Cad2 positions/sequences in cadBA promoter
- E. coli maintain a relatively constant intracellular pH (pumps sense extracellular pH)
- CysK promoter (high pH)
- luciferin regeneration
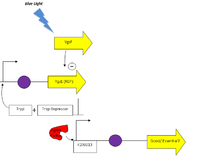
Blue Light Regulated Promoter YgcF
- Articles/ References:
- Proposed Pathway:
- Parts to Build:
- K238013
- gnl|ECOLI|G6603
- gnl|ECOLI|G6602
- Additional Parts:
- Vector: PSB1A2 (Isolated and purified from a gel)
- Insert with plac promoter and RFP gene: J04450
Maths
- Networks (Modeling Focused)
- Field might be kind of saturated; it seems like a lot of work has been done.
- But not with netLogo. How could that work?
- RePast: another ABM suite that might be mroe suited to networks [21]
- General
- Network motifs in the transcriptional regulation network of Escherichia coli
- [22] Gene Regulatory Network wikipedia page
- [23] long dissertation on modeling GRNs
- Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles
- Process Calculus
- [24] Process Calculus wikipedia page
- [BioAmbients: An abstraction for biological compartments Process Calculi for bio modelling; might be at the level of cells as opposed to genes etc.
- Boolean Networks
- [25] Boolean Network wiki page; elementary CA are special cases of Boolean networks
- Boolean modeling of GRNs
- 'The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response
- Or-Not Logic Gate with E.Coli
- Dynamical Systems
- Discrete dynamical system modelling for gene regulatory networks of 5-hydroxymethylfurfural tolerance for ethanologenic yeast
- A Linear Discrete Dynamic System Model for Temporal Gene Interaction and Regulatory Network Influence in Response to Bioethanol Conversion Inhibitor HMF for Ethanologenic Yeast
- Field might be kind of saturated; it seems like a lot of work has been done.
- Flux Balance Analysis
- Agent Based Models/Complex Adaptive Systems
- Real Computing/Complexity
- [31] Lecture transcripts from two MIT courses on compleity by a very smart guy in the field
- [32] Review of physical computing by the same researcher
- [33] Part of a textbook on computation theory
- [34] Harvard analog computing
- [35] Free draft of Princeton text on computational complexity
- [36] Paper written by one of the authors of Complexity and Real Computation that contains the same basic ideas
- [37] An analog computer museum and information site run by a Dr. Bernd Ulmann, who did his doctoral thesis on analog computing
- [38] Abstract of a 1964 study that used analog computers to model a bacterial cell
- [39] Paper on combined use of analog and digital computation
- [40] First 28 pages of Neural Networks and Analog Computing: Beyond the Turing Limit
- Neural Networks
- Fuzzy Logic/Modeling
- [44] Soft Computing (general field)
- [45] A whole book on fuzzy rule based modeling
- [46] Fuzzy modeling and control of biological processes
- Systems biology by the rules: hybrid intelligent systems for pathway modeling and discovery
- [47] FCL Java package
- [48] Noise-based logic
- [49] Fuzzy sets overview slides (MIT)
Communication
- Bacterial Conjugation
General
GGA for College Teaching Labs
6/21/12 Experiment
Objective: The goal is for the cell colonies to exhibit a visible amount of GFP in the least amount of time possible.
Process: Previously, I had seen red fluorescence in the plate of the solution that included J119022 plasmids and was in the PCR machine for 30 cycles. So, Dr. Campbell and I decided to try the same, but with 5 or 10 cycles. We also tried 5 or 10 cycles with cleaned PCR product (J23100 insert), as opposed to the entire 022 plasmid. This left us with four experimental plates and two negative controls (without enzymes; one for 5 cycles, one for 10 cycles).
Results: After letting the plates incubate overnight, we saw the results. Initially, only the 5- and 10-cycle plasmid plates produced red colonies. However, as the day went on, the PCR product plates showed fluorescence, too. The plates then sat out for the weekend, and by Tuesday morning, three of the four experimental plates were obviously red. These three were the 10-cycle 022 plasmid plate, the 10-cycle PCR product plate, and the 5-cycle PCR product plate. The 5-cycle 022 plasmid plate had a few red colonies, but not nearly as many as the other three. We then took pictures under UV light (seen below) of the three that showed fluorescence plus a negative control plate (10 cycles) as a comparison. The 5-cycle PCR product plate displays the least amount of RFP of the three shown, with roughly half of the colonies glowing red.
These inserts (J23100) were cloned into the plasmid J119044.
Figure 1. Photograph of 4 plates containing transformed E. coli (JM109) used in this GGA experiment. Clockwise from the top: Negative control, 10-cycle 022 plasmid, 10-cycle PCR product, 5-cycle PCR product
Figure 2. Photograph of the negative control plate
Figure 3. Photograph of the 10-cycle plasmid plate
Figure 4. Photograph of the 10-cycle PCR product plate
Figure 5. Photograph of the 5-cycle PCR product plate
6/28/12 Experiment
Objective: The goal of this experiment was to see if unclean PCR product worked as efficiently in Golden Gate Assembly (GGA) as the clean PCR product. If it did, this would eliminate the step of cleaning the PCR product, thereby making GGA a quicker process.
Process: I had eight experimental plates and three control plates. For the experimental plates, I made the GGA solution with different volumes of unclean PCR product instead of J119022 plasmid. I used 1, 5, 10, and 20 µL of unclean PCR product, and one of each solution went through either 5 or 10 cycles in the PCR machine.
I then compared these experimental plates to the positive control (the same solution, but with 44 ng of clean PCR product; I knew from a previous experiment that this should produce RFP) to see what volume (if any) of unclean PCR product would produce similar results to the clean PCR product. In the set of 10-cycle plates, there were two positive controls (one was 25 µL total, and the other was 10 µL; this was to see if adding more water would affect the results). Because both positive controls produced the same amount of colonies (and also the same amount of red colonies), I can conclude that the amount of water in the solutions does not affect the effectiveness of GGA.
I only had one positive control in the 5-cycle set because there was not enough clean PCR product to have two. I chose to keep the 10 µL control because I knew that it should definitely work (because of a previous successful experiment), and I didn’t know yet if the 25 µL would produce RFP (the 5-cycle plates experiment was performed a day earlier than that of the 10-cycle plates).
There was one negative control in each of the 5- and 10-cycle experiments; the solutions were the same, just without the insert (PCR product—clean or unclean).
Results: The two 10-cycle positive controls (25 µL and 10 µL) both had about 50% of their colonies glowing (see Figures blank and blank). The 5-cycle control, however, had a lawn of colonies in the middle (that did not exhibit RFP) and had red colonies on the periphery of the plate. This was confusing because last week I performed an identical experiment, and red colonies were across the entire plate, not just the periphery. I’m assuming this is a result of faulty spreading of the cells, because the colonies along the edge of the plate show a significant amount of RFP, showing that GGA was successful.
As for the experimental plates, only the 5- and 10-cycle plates containing 1 µL of unclean PCR product showed RFP, with the 10-cycle plate showing even more (see Figures blank and blank). The plates containing 5, 10, and 20 µL of unclean PCR product did not exhibit RFP at all in either the 5- or 10-cycle plates, and in fact the amount of colonies on the plates had a negative correlation with how much unclean PCR product was added. I assume that, because the positive controls are already glowing so brightly by this point, all of the plates would have exhibited some red by now if they were going to. Thus, the plates that don’t have any red (the 5- and 10-cycle plates of 5, 10, and 20 µL unclean PCR prodcut, and the negative controls) probably will not ever exhibit RFP if they have not already. Thus, because the 1 µL solutions showed RFP, I decided to go forward with these.
I plan to ligate and transform solutions equal to those I used for the plates with 1 µL of unclean PCR product, except they will be in the PCR machine for 20 or 30 cycles. Because the 10-cycle plate with 1 µL of unclean PCR product produced more RFP than the 5-cycle plate of the same solution, I am guessing that the more cycles the solution goes through, the higher percentage of colonies will exhibit RFP.
If this is the case, then professors can have options of how to perform GGA in their classroom. They can choose to add the extra step of cleaning the PCR product (and then only have to put the solution in for 5 cycles in the PCR machine), or they can not clean the PCR product and put in the PCR machine for more time (if this ends up being a viable option; I will find out soon).
These inserts (J23100) were cloned into the plasmid J119044.
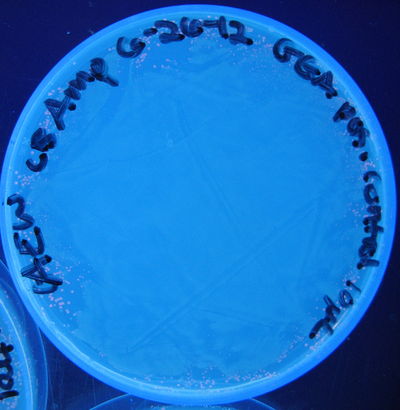
Figure 1: Photograph of a plate containing transformed E. coli (JM109) used in this GGA experiment; Positive control, 5 cycles.
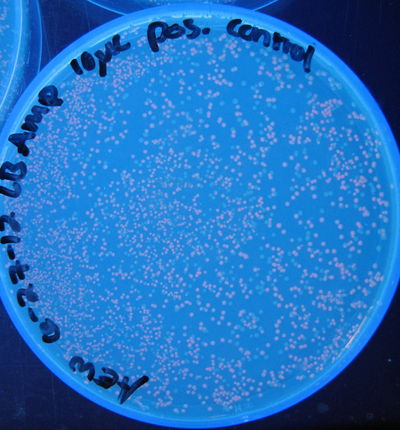
Figure 2: Photograph of positive control plate, 10 µL, 10 cycles
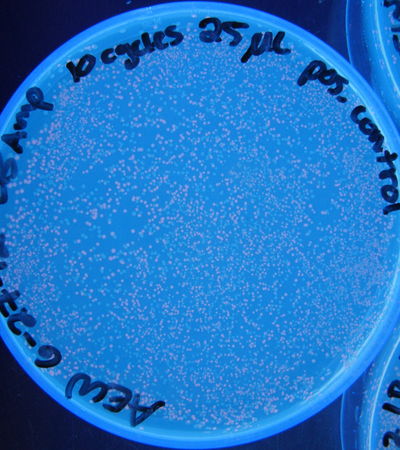
Figure 3: Photograph of positive control plate, 25 µL, 10 cycles

Figure 4: Photograph of 1 µL unclean PCR product plate, 5 cycles
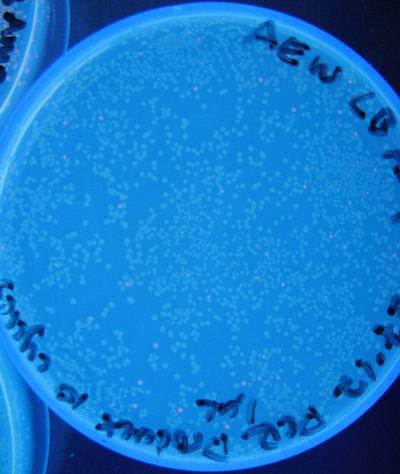
Figure 5: Photograph of 1 µL unclean PCR product plate, 10 cycles
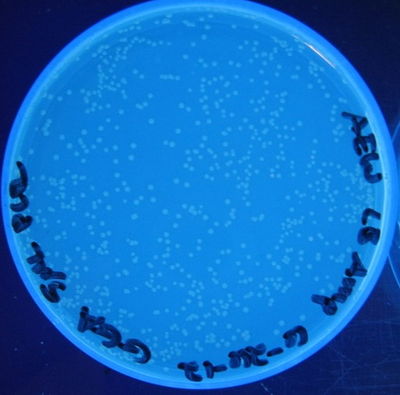
Figure 6: Photograph of 5 µL unclean PCR product plate, 5 cycles