Difference between revisions of "Hypothetical Protein 644029933 (Olivia Ho-Shing)"
| (One intermediate revision by the same user not shown) | |||
| Line 6: | Line 6: | ||
#[http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm Kyte-Doolittle Plot] | #[http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm Kyte-Doolittle Plot] | ||
#[http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html PREDATOR] | #[http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_preda.html PREDATOR] | ||
| + | #Search for Shine Dalgarno sequence within 50 bp upstream | ||
== BLASTn and BLASTp == | == BLASTn and BLASTp == | ||
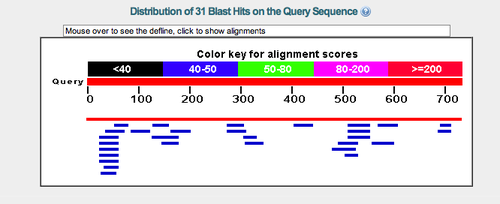
The nucleotide sequence for this gene does have a start codon (ATG) and stop codon (TGA) defined. The BLASTn hits for the nucleotide sequence, other than a perfect alignment with itself (Halomicrobium mukohataei complete genome), returned hits with poor Query coverage (<10%) and unreliable E-values (>0.05). <br> | The nucleotide sequence for this gene does have a start codon (ATG) and stop codon (TGA) defined. The BLASTn hits for the nucleotide sequence, other than a perfect alignment with itself (Halomicrobium mukohataei complete genome), returned hits with poor Query coverage (<10%) and unreliable E-values (>0.05). <br> | ||
| − | [[Image:hyp-blastn|500px|center]]<br> | + | [[Image:hyp-blastn.png|500px|center]]<br> |
| + | |||
| + | For the BLASTp hits, there were several hits for a monooxygenase protein, but the E-values were very unreliable. | ||
| + | <br>[[Image:hyp-blastp.png|500px|center]]<br> | ||
| + | [[Image:hyp-blastp-align.png|thumb|500px|center]]<br> | ||
| + | |||
| + | == Kyte-Doolittle Plot == | ||
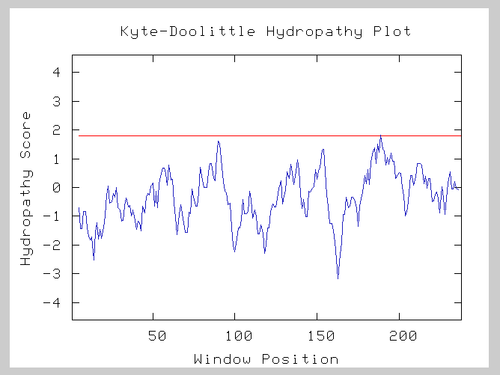
| + | Kyte Doolittle hydropathy plots use an amino acid sequence to predict whether the protein crosses the plasma membrane, as in an integral membrane protein. Predicting if this hypothetical protein is an integral membrane protein would give some clue to its function. When I submitted the amino acid sequence, the plot indicated that this sequence probably does not cross the plasma membrane (If it did, there would be peaks spanning above the red line at 1.8). | ||
| + | <br>[[Image:kyte-doolittle.png|500px|center]]<br> | ||
| + | |||
| + | == PREDATOR == | ||
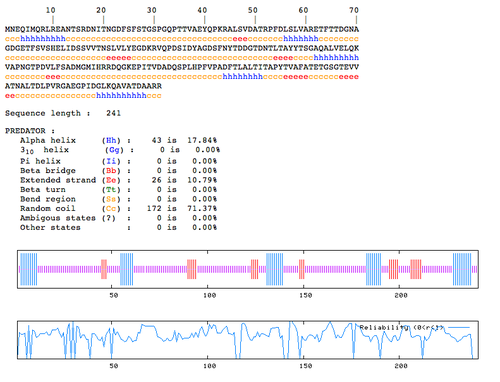
| + | PREDATOR predicts the 3D structure of an amino acid sequence, which could also help in predicting the function. Here are the results I got from submitting the hypothetical protein sequence: | ||
| + | <br>[[Image:hyp-predator.png|500px|thumb|center]]<br> | ||
| + | |||
| + | I also wanted to see what results I got from a [http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=protein&dopt=GenPept&RID=AFEVZ8ST012&log%24=prottop&blast_rank=6&list_uids=167567057 monooxygenase] from the BLASTp hits: | ||
| + | <br>[[Image:mono-predator.png|500px|thumb|center]]<br> | ||
| + | |||
| + | The hypothetical protein looks like it could be similar to a portion of the monooxygenase, or it could be something completely different. | ||
| + | |||
| + | == Shine Dalgarno == | ||
| + | The 50 bp upstream of the start codon are: TCTGGAGATCGACCGCAAGGCCGTGACCGTCCGAGCGGAGGACTGACAGC | ||
| + | I found what looks like a Shine Dalgarno sequence (CGGAGGA) 9 bp upstream of the start codon, which around the average separator length for our genome. | ||
Latest revision as of 14:59, 9 September 2009
I randomly chose a protein with no predicted function: 644029933 hypothetical protein
In order to search for some possible function I tried using:
- BLASTn
- BLASTp
- Kyte-Doolittle Plot
- PREDATOR
- Search for Shine Dalgarno sequence within 50 bp upstream
BLASTn and BLASTp
The nucleotide sequence for this gene does have a start codon (ATG) and stop codon (TGA) defined. The BLASTn hits for the nucleotide sequence, other than a perfect alignment with itself (Halomicrobium mukohataei complete genome), returned hits with poor Query coverage (<10%) and unreliable E-values (>0.05).
For the BLASTp hits, there were several hits for a monooxygenase protein, but the E-values were very unreliable.
Kyte-Doolittle Plot
Kyte Doolittle hydropathy plots use an amino acid sequence to predict whether the protein crosses the plasma membrane, as in an integral membrane protein. Predicting if this hypothetical protein is an integral membrane protein would give some clue to its function. When I submitted the amino acid sequence, the plot indicated that this sequence probably does not cross the plasma membrane (If it did, there would be peaks spanning above the red line at 1.8).
PREDATOR
PREDATOR predicts the 3D structure of an amino acid sequence, which could also help in predicting the function. Here are the results I got from submitting the hypothetical protein sequence:
I also wanted to see what results I got from a monooxygenase from the BLASTp hits:
The hypothetical protein looks like it could be similar to a portion of the monooxygenase, or it could be something completely different.
Shine Dalgarno
The 50 bp upstream of the start codon are: TCTGGAGATCGACCGCAAGGCCGTGACCGTCCGAGCGGAGGACTGACAGC I found what looks like a Shine Dalgarno sequence (CGGAGGA) 9 bp upstream of the start codon, which around the average separator length for our genome.