Difference between revisions of "Genomic Insertion Protocol"
Wideloache (talk | contribs) |
Wideloache (talk | contribs) |
||
| Line 1: | Line 1: | ||
*Warning: This protocol needs to be modified an updated based on the results obtained in my first attempt at genomic integration. Please see my [http://gcat.davidson.edu/GcatWiki/index.php/Will_DeLoache_Notebook lab notebook results] until then.*** | *Warning: This protocol needs to be modified an updated based on the results obtained in my first attempt at genomic integration. Please see my [http://gcat.davidson.edu/GcatWiki/index.php/Will_DeLoache_Notebook lab notebook results] until then.*** | ||
| + | |||
| + | |||
| + | This genomic insertion protocol utilizes [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=100134 CRIM technology] and was modified by the [http://andersonlab.qb3.berkeley.edu/ Anderson Lab at UC Berkeley]. For more information on the technology, you can read the [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=100134 article on CRIM technology]. | ||
| + | |||
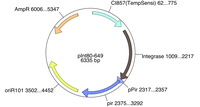
| + | You will need two plasmids: pG80ko and pInt80-649. pG80ko can be found in the [http://gcat.davidson.edu/GCATalog-r1/Login.php freezer stocks] in box 4-1, position 55. It contains a stuffer insert that is about 700 bps and should be replaced with the part you want to insert. pInt80-649 can be found in the freezer stocks in box 4-1, position 51. pG80ko's sequence can be found [http://gcat.davidson.edu/GcatWiki/images/b/bb/PG80ko.doc here]. pIn80-649's sequence can be found [http://gcat.davidson.edu/GcatWiki/images/e/ec/PInt80-649.doc here]. | ||
| + | |||
| + | This plasmid maps are shown below: | ||
| + | |||
| + | [[Image:PG80ko.jpg|200px|thumb|none|pG80ko containing BBa_K091206 for insertion. This is where your part will be.]] [[Image:PInt80.png|200px|thumb|none|pInt80-649.]] | ||
| + | |||
| + | |||
Revision as of 13:46, 15 May 2009
- Warning: This protocol needs to be modified an updated based on the results obtained in my first attempt at genomic integration. Please see my lab notebook results until then.***
This genomic insertion protocol utilizes CRIM technology and was modified by the Anderson Lab at UC Berkeley. For more information on the technology, you can read the article on CRIM technology.
You will need two plasmids: pG80ko and pInt80-649. pG80ko can be found in the freezer stocks in box 4-1, position 55. It contains a stuffer insert that is about 700 bps and should be replaced with the part you want to insert. pInt80-649 can be found in the freezer stocks in box 4-1, position 51. pG80ko's sequence can be found here. pIn80-649's sequence can be found here.
This plasmid maps are shown below:
Note that you'll need a pir strain for replication of pG80 plasmids. You can drop your biobrick into the Eco/pst sites of pG80ko, transform the pir116 cells. Grow up, miniprep, and map a single colony. Make competent cells of pInt80-649 in your target strain (plate them on Amp). It is temperature sensitive, so do all growth manipulations at 30 degrees. Transform in your pG80 derivative, plate on 15ug/mL gentamicin plates at 37 degrees. Grow a single colony to saturation at 37 in LB+15ug/mL gentamicin, then restreak on a gentamicin plate at 43 degrees. You can use the oligos below to PCR amplify the phi80 locus for confirmation of integration and sequencing.
attPhi80-1: CTGCTTGTGGTGGTGAAT
attPhi80-2: ACTTAACGGCTGACATGG
attPhi80-3: ACGAGTATCGAGATGGCA
attPhi80-4: TAAGGCAAGACGATCAGG
| sequence Temp (°C) | No integrant with 1 and 4 | Single integrant with 1 and 2, 3 and 4 | Multiple integrant with 1 and 2, 3 and 2, 3 and 4 |
| 63 | 546 | 409, 732 | 409, 595, 732 |