Difference between revisions of "Davidson College"
Macampbell (talk | contribs) |
Macampbell (talk | contribs) (→Davdison College has experimented with different uses for DNA microarrays.) |
||
| Line 4: | Line 4: | ||
CGH means you isolate total genomic DNA from two strains, label them different colors, and hybridize them to a normal yeast (that's what we use) DNA microarray. If the yeast has experience aneuploidy, then the genes that are duplicated or deleted will display green or red. Genes that are present at the same level in both strain will appear yellow. | CGH means you isolate total genomic DNA from two strains, label them different colors, and hybridize them to a normal yeast (that's what we use) DNA microarray. If the yeast has experience aneuploidy, then the genes that are duplicated or deleted will display green or red. Genes that are present at the same level in both strain will appear yellow. | ||
[http://www.bio.davidson.edu/projects/GCAT/CGH/CGH.html You can access the protocol from the GCAT "CGH" web page.] | [http://www.bio.davidson.edu/projects/GCAT/CGH/CGH.html You can access the protocol from the GCAT "CGH" web page.] | ||
| + | <br> | ||
| + | |||
| + | [[Image:CGHNov27.jpg|429px|]] | ||
| + | Whole yeast genome microarray probed with <italics>wild-type</italics> yeast gDNA for both channels (Cy3 adn Cy5). | ||
| + | <br> | ||
| + | <hr> | ||
<br> | <br> | ||
Revision as of 20:12, 27 February 2006
Davdison College has experimented with different uses for DNA microarrays.
- We have developed protocols for comparative genome hybridization (CGH).
CGH means you isolate total genomic DNA from two strains, label them different colors, and hybridize them to a normal yeast (that's what we use) DNA microarray. If the yeast has experience aneuploidy, then the genes that are duplicated or deleted will display green or red. Genes that are present at the same level in both strain will appear yellow.
You can access the protocol from the GCAT "CGH" web page.
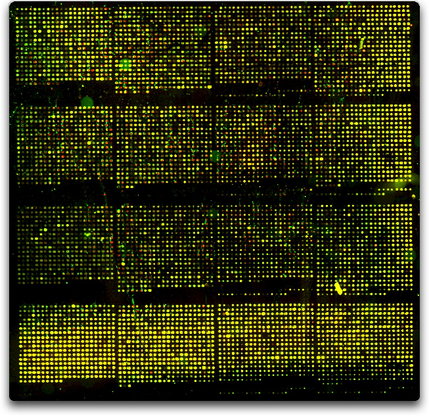 Whole yeast genome microarray probed with <italics>wild-type</italics> yeast gDNA for both channels (Cy3 adn Cy5).
Whole yeast genome microarray probed with <italics>wild-type</italics> yeast gDNA for both channels (Cy3 adn Cy5).
- We have also developed teaching chips that allow any school to print their own chips and do controlled experiments using oligos as probes.
You can use any piece of DNA as the target, but we have found cloned PCR products work especially well. By using PCR products, you can target any gene or cDNA. Ideally, you will want to balance the GC content and size of the PCR products. Furthermore, but cloning the PCR products into a plasmid, you can produce very large quantities of target DNA for very little expense. You can also use empty plasmid as negative control DNA.
