Difference between revisions of "Will Notebook1"
Wideloache (talk | contribs) (→Wideloache 18:06, 20 October 2008 (EDT)) |
Wideloache (talk | contribs) |
||
| Line 15: | Line 15: | ||
I performed a ligation with 1uL of vector and 3uL of insert (and a negative control). I then did a Zyppy transformation and plated on Amp plates. | I performed a ligation with 1uL of vector and 3uL of insert (and a negative control). I then did a Zyppy transformation and plated on Amp plates. | ||
=== === | === === | ||
| − | + | <pre> | |
| + | Begin making competent cells for genomic insertion. | ||
| + | </pre> | ||
==[[User:Wideloache|Wideloache]] 23:56, 19 October 2008 (EDT)== | ==[[User:Wideloache|Wideloache]] 23:56, 19 October 2008 (EDT)== | ||
On Thursday night I didn't leave the shaker on, so on Friday I turned the shaker on for an extra 5-6 hours and put my 6 cultures in the fridge. Today I miniprepped the 6 cultures. I plan to build the part Prom-LuxR-TT-Prom-LasR-TT. I will use K091204 as a back vector and K091205 as a back insert. In order to verify K091204, I also digested each K091204 MP with EcoRI/PstI. All digestions were done with 5 uL of DNA. | On Thursday night I didn't leave the shaker on, so on Friday I turned the shaker on for an extra 5-6 hours and put my 6 cultures in the fridge. Today I miniprepped the 6 cultures. I plan to build the part Prom-LuxR-TT-Prom-LasR-TT. I will use K091204 as a back vector and K091205 as a back insert. In order to verify K091204, I also digested each K091204 MP with EcoRI/PstI. All digestions were done with 5 uL of DNA. | ||
Revision as of 22:08, 20 October 2008
Wideloache 18:06, 20 October 2008 (EDT)
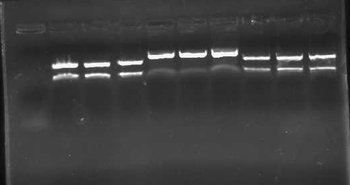
Today I ran last night's digestions on a .8% gel and found that all 3 clones for each construct appeared to be correct. Unfortunately, our ladder went bad at some point, so the ladder didn't work (it has now been replaced). Luckily, I was looking for an insert size of ~1000bps for a successful ligation. An unsuccessful ligation would have given an insert size of ~50bps. I gel purified samples K091204-B (S/P) and K091205-B (X/P) for use in the next step of assembly.
I performed a ligation with 1uL of vector and 3uL of insert (and a negative control). I then did a Zyppy transformation and plated on Amp plates.
Begin making competent cells for genomic insertion.
Wideloache 23:56, 19 October 2008 (EDT)
On Thursday night I didn't leave the shaker on, so on Friday I turned the shaker on for an extra 5-6 hours and put my 6 cultures in the fridge. Today I miniprepped the 6 cultures. I plan to build the part Prom-LuxR-TT-Prom-LasR-TT. I will use K091204 as a back vector and K091205 as a back insert. In order to verify K091204, I also digested each K091204 MP with EcoRI/PstI. All digestions were done with 5 uL of DNA.
Streak successful assemblies of K091204 and K091205
Wideloache 14:09, 16 October 2008 (EDT)
I added 2 parts to the registry today:
K091204 = J23100 + I0462
K091205 = J23100 + S03954
I picked 3 clones from each of the transformation plates and grew them up O/N
Make plates with varying Amp and agar Concentrations. Prepare to develop mathematical model of amp growth.
Wideloache 14:57, 9 October 2008 (EDT)
I ran my digestion of the 3 LuxR/LasR cassette on a .8% gel. I excised the large band from Lane 1 and the small bands from Lanes 2 and 3.
I did gel purification of these parts and ligated using 5uL of DNA:
J23100+I0462
J23100+S03954
J23100 (neg control)
I finally did a zyppy transformation and plated on Amp plates for O/N growth.
Wideloache 20:04, 8 October 2008 (EDT)
I finally finished making a time-lapse video of colony growth using beta-lactamase diffusion to create time delayed cell growth. The video can be viewed here. The method appeared to work as we had hoped. In the top right corner of the video, I spotted ampicillin resistant cells. I think spotted non-amp resistant cells along the horizontal and diagonal axes and streaked them along the vertical axis. Cell growth was delayed in colonies that were farther from the amp resistant colony, suggesting that beta lactamase was diffusing through the agar.
Wideloache 15:55, 7 October 2008 (EDT)
Because the primer dimer assembly method was continuing to cause mutations, even with the modified protocol, I decided to move on to the oligo assembly method for both pLux' and pLuxLas. I used the Lancelator program to design 4 oligos per promoter. These oligos were built to assemble with with EcoRI and PstI stick ends flanking the DNA segment. Two of the 4 oligos could be reused in both promoter assemblies, so the total oligo order was 6. Go here to download my oligo design file.
I also began working on the LuxR/Lasr cassette again. Since S03983 (J23100-LuxR) had been the wrong part, I now needed a new source of LuxR. I found a registry part, BBa_I0462 (LuxR-Terminator) that would be a useful intermediate in the construction of J23100-LuxR-Terminator. I looked back over the iGEM team's lab notebooks and found that they had encountered difficulties when trying to take this part out of the registry. Instead, they had made the part from its two basic parts. I tried to find a miniprep or freezer stock of BBa_I0462, but had trouble at first. Then I realized that Erin had mislabeled some of her minipreps (calling I0462 I0426 instead). I verified the date of the miniprep in her notebook (7/21/2008) and decided to use that prep for my subsequent construction. I will now proceed by putting a promoter in front of both I0462 and S03954 (colonies that are unsuccessful show fluoresce red). I will then put those two parts together and do the genomic insertion.
Today I began the following O/N digestions:
J23100 - Back Vector (S/P)
S03954 - Back Insert (X/P)
I0462 - Back Insert (X/P)
5 uL DNA
11 uL H20
2 uL Buffer
1 uL RE #1
1 uL RE #2
Remove S03983 (and possibly a mislabeled I0426) from the -80 Freeze down any parts that are not frozen down: pir+ strain, J23100, S03954, B0015, I0462, pG80k0, pInt80-649
Wideloache 18:45, 6 October 2008 (EDT)
A lot of my lab time in the past week has been devoted to making a video file of the time-delayed growth system using beta-lactamase. I have devoted time to this while I awaited sequencing results from Clemson. I had to resend all 8 samples because they lost all of Davidson's sequencing samples. I finally got the results back today. I found mutations in all 3 pLuxLas samples that I submitted for sequencing. I have included a multiple alignment below that shows where the mutations were in the various clones. Clone 1 had a single deletion in the -10 region. Clones 2 and 3 had multiple mutations. No mutation appeared across all 3 clones, suggesting that each was a random mutation that most likely results from PCR amplification.
Expected GAATTCGCGGCCGCTTCTAGAGACCTGTAGGATCGTACAGGTTGACATCTATCTCATTTGCTAGTATAATCGAATAAATACTAGTAGCGGCCGC 94
1 GAATTCGCGGCCGCTTCTAGAGACTTGTAGGATCGTACAGGTTGACATCTATCTCATTTGCTAG-ATAATCGAATAAATACTAGTAGCGGCCGC 93
2 GAATTCGCGGCCGCTTCTAGAGACCTGTAGGATCGTACAGGTTGACATCTATCTCATTTGCTAGTATA-TCGAATAAA-ACTAGTAGCGGCCG- 91
3 GAATTCGCGGCCGCTTCTAGAGACCTGTAGGATCGTACAGGTTGACATCTATCTCATTTGCTAGTAAA-TCGAATAAATACTAGTAGCGGCCG- 92
************************ *************************************** * * ********* **************
I also got results back for the sequencing of the LuxR/LasR cassette. This confirmed that B0015, J23100, and S03954 were all correct. However, S03983 appeared to be J23100-LasR (S03982) instead of J23100-LuxR (S03983). The LasR contained in the part also had a nonsense mutation in it. I will have to find the LuxR basic part to use in further assemblies. I also need to remove part S03983 from the -80, since it is incorrect.
Wideloache 16:00, 29 September 2008 (EDT)
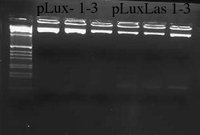
I ran last night's digestion of pLux and pLuxLas clones on a 2.2% gel. Band size was expeced to be 95 bps for each clone. pLuxLas clones appeared correct and were sent off for sequencing. pLux clones did not appear to have the correct insert size and will have to be re-cloned.
I sent the 3 pLuxLas clones off to be sequenced.
Wideloache 20:38, 28 September 2008 (EDT)
I did a Promega miniprep on the 6 colonies that I had picked on Friday. I digested each of these minipreps with EcoRI/PstI. If I get an insert of ~55 bps then I will send the samples off for sequencing.
Run the 6 digestions on a gel
Wideloache 13:56, 26 September 2008 (EDT)
For the transformations of the pLux and pLuxLas promoters I had significant growth. I had no colonies on the negative control transformation of pSB1A2. Many colonies on both the pLux and pLuxLas plates were red because an RFP insert had been digested out of the pSB1A2 plasmid. I picked 3 non-red colonies from each stock and grew them up in LB+Amp for miniprepping tomorrow.
Wideloache 13:59, 25 September 2008 (EDT)
Today I began the construction of the two Lux promoters - pLux and pLuxLas. These promoters will be constructed using primer extension. The primers used overlapped 20 bps on the 3' end. I used the following PCR protocol to extend the primers:
1 ug of primer 1
1 ug of primer 2
18 ul of H20
20 ul of Monster Mix
PCR Program = PrimerEx:
- 5 mins at 94 C
- 30 secs at 94 C
- 15 secs at 55 C
- 30 secs at 74 C
- Repeat steps 2-4 for 4X
- 5 mins at 74 C
- Infinity at 21 C
I then did a Zymo cleanup of the PCR reaction and and eluted into 10 uL of H20. I then digested the two PCR extensions with EcoRI and PstI:
5 uL of DNA
11 uL of H20
2 uL of Buffer H
1 uL of EcoRI
1 uL of PstI
I did another Zymo cleanup to remove the enzymes from digestions before ligating and transforming. I ligated with pSB1A2 plasmid from the iGEM gel purification box. I transformed into JM109 cells (Zyppy transformation).
Wideloache 14:39, 23 September 2008 (EDT)
I ran the digestions on a .8% gel, and obtained the following result:
At this point I realized that I had a couple of problems:
Problem 1: At some point along the way, I switched the Registry #'s of S03954 (LasR-TT) and S03983 (Prom-LuxR) with their descriptions. Therefore, I had done the incorrect digestion for each of these parts. S03954 needs to be digested as a back insert (and then ligated with J23100) while S03983 needs to be digested as a front insert (and then ligated with B0015).
Problem 2: The J23100 part that I digested with S/P gave an unexpected band at ~1000bp. I went back and looked at the documentation for this part and found that in the registry, it exists in the plasmid J61002. This plasmid has RFP between the Spe and Pst sites. I need to determine if the part was moved before ligating it with LuxR and other parts. If so, I need to locate this sample or else move it again myself into pSB1A2. If not, then I may have to redo the assembly of S03983.
After talking with Dr. Campbell, it appeared likely that the J23100 was indeed in the incorrect plasmid (J61002). I decided to sequence the 4 different parts that I was assembling to confirm what I had. I sequenced S03954 and S03983 with VR. I also sequenced S03983, B0015, and J23100 with VF2. While I wait for the results of that sequencing i will go ahead with primer dimer assembly.
make freezer stocks of: S03954 J23100 B0015 Ec100D::pir116 cells from Chris
Wideloache 13:58, 22 September 2008 (EDT)
Last night I grew up S03983 (LasR-TT) in LB. Today I did a Zyppy MP and obtained a concentration of 139.9 ng/uL. I then performed the following digestions:
13 uL H20
3 uL DNA
2 uL Buffer
1 uL RE 1
1 uL RE 2
S03954 -> Front Insert (E/S) + B0015 -> Front Vector (E/X)
J23100 -> Back Vector (S/P) + S03983 -> Back Insert (X/P)
This is the first of two rounds of ligation need to make the construct: Prom-LuxR-TT-Prom-LasR-TT. This will be inserted into the genome using the plasmids shipped from the Anderson Lab last week.
Wideloache 14:44, 19 September 2008 (EDT)
I obtained minipreps of B0015, J23100, and S03954 (Promoter-LuxR) from the iGEM racks. I couldn't find a miniprep of S03983 (LasR-TT), but I got a sample from the -80. Note: S03954 was not in the -80 as far as I could tell. I will freeze this down along with the other MP's after I transform them into cells.
I also received the shipment from Chris and my oligos were shipped today. I will begin with pLas assembly and LuxR/LasR genome insertions this weekend.
Wideloache 14:03, 18 September 2008 (EDT)
Over the past few days, I have been getting up to speed on the project as a whole. A lot of time was spent figuring out the primer sequences for the pLux' and pLuxLas promoters. I found that the primers that had been ordered over the summer used the -10 region from the registry, which differed from the standard consensus sequence in E. coli. I also found that a tube switch had occurred over the summer since the sequences for pLux' matched the expected sequence for pLuxLas and vice versa. Still, all 6 clones had at least one mutation in them. I decided to order new primers with the consensus -10 region. This only required that I reorder the reverse primers. I will reuse the forward primers that were designed over the summer.
I also contact Chris Anderson about performing a genomic insertion of LuxR and LasR. He is in the process of shipping the necessary plasmids to me. I will be doing a phi80 genome integration (protocol).
Wideloache 16:20, 10 September 2008 (EDT)
On Monday, I got K091136, K091146, and K091117 out of the -80. I need to make pLux' and pLux+Las- and do a genomic insertion of LuxR/LasR before I can test the XOR promoters.
Begin Primer Synthesis of pLux' and pLux+Las- (could consider site directed mutagensis with Pallavi's primers) Email Chris about genomic insert vector Talk to Erin Feeney about the status of the LuxR and LasR cassettes