Modeling Promoter Activity
Contents
Modeling Promoter Activity
In order to use synthetic promoters to their fullest potential, we have to understand how they work. Sythetic promoters cannot help us model gene circuit activity unless models are developed for the activity of the promoter itself. Determining how exactly a promoter's strength correlates to its mutations is not easy, since for the most part it requires working with promoters on the level of individual sets of nucleiotides.
Jensen and Hammer (1997): Spacer Sequences
In this 1997 paper, Jensen and Hammer constructed a library of synthetic promoters based on the Lactococcus lactis prokaryotic promoter in order to better determine how gene sequence of promoters was tied to the promoter strength. Specifically, Jensen and Hammer were looking for a way to construct a constitutively active promoter – one that was always turned on, without needing an inducer – that could be safely used to tune gene expression in industrial-scale metabolic engineering projects, where inducers might be impractical or hazardous.
In order to tune the steady-state L. lactis promoter without using an inducer, Jensen and Hammer had to create a library of L. lactis mutant promoters, all with various levels of activity. To generate the library, they used the method described in Promoters and Reporters in Synthetic Biology: constructing oligonucleiotides that matched the genes common to all previous L. lactis promoters and mutants, then allowing the oligonucleiotides to be joined together by random spacer sequences.
After the promoter library was synthesized, promoters were cloned into both L. lactis and E. coli; each cell culture containing a different promoter was tested for the level of beta-galactosidase activity. The activity of each promoter (in Miller units, or beta-galactosidase concentration) is described in Figure 3.
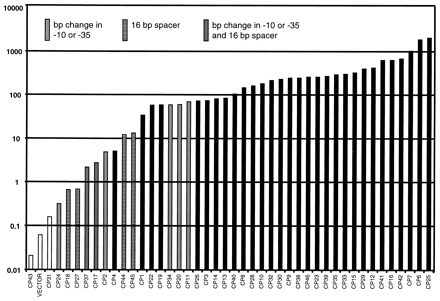 Figure 3. Library of synthetic promoters for L. lactis. Promoter activities (Miller units) were assayed from the expression of a reporter gene (lacLM) encoding -galactosidase transcribed from the different synthetic promoter clones on the promoter cloning vector pAK80. The patterns of the data points indicate which promoter clones contain errors in either the 35 or the 10 consensus sequence or in the length of the spacer between these sequences. From Jensen and Hammer (1997). Permission Pending.
Figure 3. Library of synthetic promoters for L. lactis. Promoter activities (Miller units) were assayed from the expression of a reporter gene (lacLM) encoding -galactosidase transcribed from the different synthetic promoter clones on the promoter cloning vector pAK80. The patterns of the data points indicate which promoter clones contain errors in either the 35 or the 10 consensus sequence or in the length of the spacer between these sequences. From Jensen and Hammer (1997). Permission Pending.
The mutant promoters expressed a wide range of activity, increasing in small increments. Note that not all of the clones were "perfect" - a few had mutations in the oligonucleotide sequences that were supposed to be preserved across the library. Those clones are indicated in the graph above. However, their data was not removed because it was within range of the data from the perfect clones - they caused no break in the general data trend. In addition, all clones were tested to ensure that they were truly constitutive.
When the promoters were cloned into E. coli, the same basic trend was observed. While the promoters did not demonstrate the same level of activity as they did in L. lactis, there was still a wide range of activity observed, with the activity level increasing in steady increments.
Jensen and Hammer constructed a library of synthetic promoters that could be constitutively expressed and covered a range of activity levels, but it was still not known for certain what caused a certain promoter to be active at a certain rate. Jensen and Hammer suggested in their Discussion that "it seems that the overall three-dimensional structure which arises from a particular nucleiotide sequence could be important".
Jensen, Alper, Fischer, and Stephanopoulis (2006): Statistical Modeling and Critical Mutation Sites
In this paper, Jensen et al tried to determine exactly why some promoters in a promoter library were stronger than others, and which mutations might cause the change in strength. Jensen et al propose to examine promoter libraries statistically rather than via assays; they will determine which mutations are associated with which phenotypes based on when they appear.
Say, for example, that you are creating a mutant library of a protein that can fluoresce one of three colors: red, blue, or green. If a given point mutation – let’s call it A – has no effect on the color of the fluorescence, then (assuming the mutagenesis is truly random) that mutation should appear in every phenotype proportional to the amount of protein with that phenotype. It will not appear in one phenotype significantly more than the others unless there is significantly more protein with that phenotype. It follows, then, that if point mutation B appears much more often in, say, blue protein without there being much more blue protein than red or green protein, mutation B might have some effect on the protein’s phenotype. It is probably not the sole cause of the blue color, but it is associated with it.