Olivia's Assignment
I want to explore the question Does high salinity affect the proton pump? So I'd like to compare the rhodopsin hydrogen pump gene(s) found in H. mukohataei to those found in other halophiles and non-halophiles. Goals/To Do List:
- Find opsin gene(s) for H. mukohataei and other related halophiles
- Find opsin genes in other archaea
- BLASTp and BLASTn
- Compare GC content - does it look like the rest of the host genome
- Make phylogenetic tree based on opsin
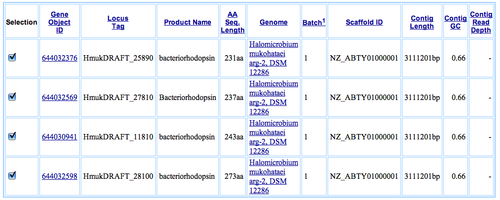
Using JGI, I found these known protein sequences predicted to be the proton pump in H. mukohataei:
- 644030941 bacteriorhodopsin
- 644032376 bacteriorhodopsin
- 644032569 Bacteriorhodopsin
- 644032598 bacteriorhodopsin
[[1]]
Other Species found to have Bacteriorhodopsin
- Natronomonas pharaonis
- H. vallismortis (ATCC 29715) sopII and htrII genes for sensory rhodopsin II apoprotein and sensory rhodopsin II transducer ACCESSION Z35308
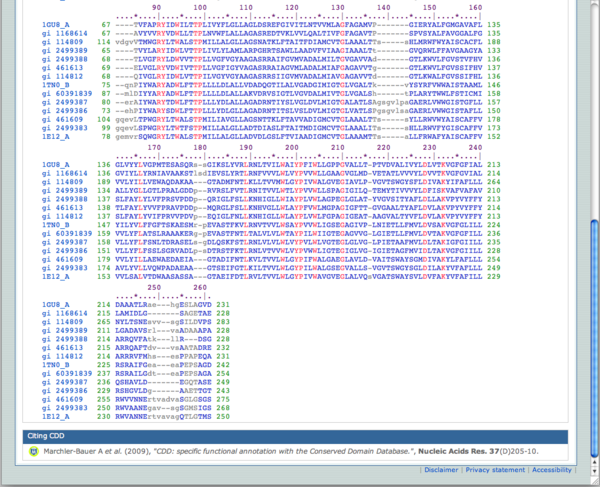
I searched the Conserved Domains Database for pFam01036 bacteriorhodopsin. I aligned all 14 rows to compare the bacteriorhodopsin sequences. From this information, I can see the generall diversity within the bacteriorhodopsin gene and identify the species that have the gene. I can then BLAST our genome's bacteriorhodopsin genes to find more dissimilar species - non-halophiles - to compare in a phylogenetic tree.