Determining whether genes called in JGI and RAST are identical
When working with multiple annotations, it is often useful to determine if a gene called by one annotation service is called by the other annotation service. The easiest way to determine if genes are called in both annotations is to compare start and stop codons. For large numbers of genes, Perl programs can be created to automate the process of comparing annotated genomes. However, if only a small number of genes are being compared, it can often be more efficient to compare start and stop codons manually.
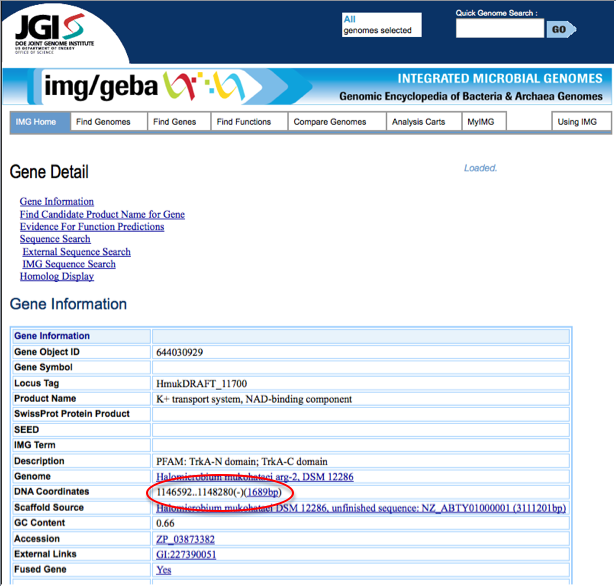
To compare start and stop codons from JGI and RAST, first one must find the start and stop codons for the gene of interest. In JGI, start from the [Gene detail page]. On this page, under Gene Information, the start and stop codon are listed next to DNA coordinate. JGI also labels whether the gene is on the plus strand or minus strand of DNA. The start and stop codons for a sample gene from JGI are circled in red below.
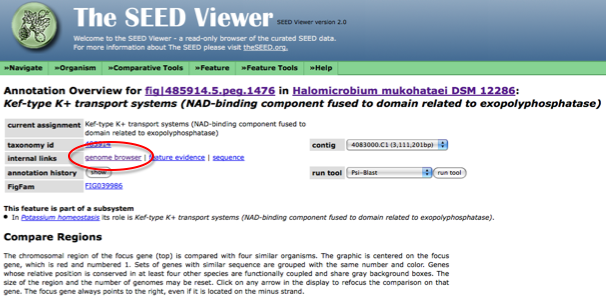
To find the start and stop codon for a gene in RAST, first browse the genome using the SEED viewer. Once you have selected your gene of interest, go to it's [Annotation overview page]. Once on this page, next to internal links select the genome browser (see figure below).
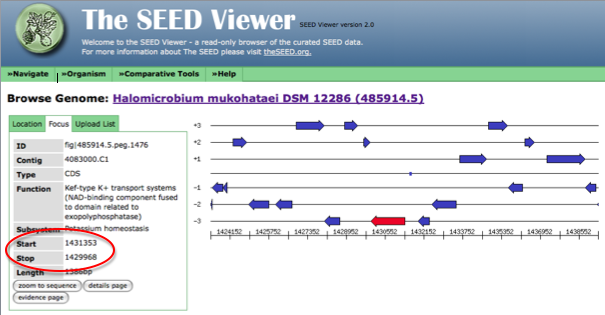
After clicking on the genome browser link, the stop and start codon for you gene will be displayed as shown in the figure below.