M13 Bacteriophage Production Protocol
This protocol describes how to produce plaques using M13 bacteriophage, and how to then harvest phage from these plaques. This is a multistep process which requires making both bottom agar (a lower layer of agar with antibiotics) and top agar (a top layer of agar with MgCl2), as well as growing host cells and performing phage dilutions.
Contents
Preparing Phage
1. Grow 3mL of cells with the phage plasmid in liquid media overnight. These cells produce M13 phage which can then be isolated from the liquid media. For example, grow J100265 + J100270 S1030 cells in LB + 50ug/mL carbenicillin + 50ug/mL kanamycin to produce SPT7-spec phage.
2. Centrifuge 1.5mL of cells at full speed (~15,500 RPM) for at least two minutes. The cells will be spun down into a pellet, while the phage will remain in the supernatant. Save the supernatant in a separate microfuge tube and discard the tube with the pelleted cells. The phage will remain viable for weeks if stored at 4°C.
3. When preparing to plate phage, make serial dilutions of the phage stock in LB broth. Each dilution should have a final volume of >10uL. Create a broad range of dilutions for plating, as too high of a phage concentration will produce many small, crowded plaques, while too low of a concentration may not produce plaques. Generally, a range from 10^-3 to 10^-9 should be sufficient.
Preparing Top and Bottom Agar
1. Bottom agar should be produced following the same recipe for creating plates for plating cells. Plates with the appropriate antibiotics that were already made for other purposes are acceptable. There’s no need to pour plates with a thinner layer of agar. Bottom agar should have the antibiotics that the host cells are resistant to, but not the antibiotics that the phage genome creates resistance for. For example, if using J100265 cells, bottom agar should have 50ug/mL carbenicillin, but not kanamycin. If host cells are S1030s, bottom agar should also have 10ug/mL tetracycline to select for the F’ plasmid.
2. Make top agar with 7g/L LB agar, 20g/L LB broth, and 5 mM MgCl2. Autoclave top agar, then transfer in 3mL aliquots to sterile tubes. Incubate in a water bath at 47°C until use. Make certain that the top agar has cooled to 47°C if it’s being used immediately after autoclaving. If top agar is too hot, it will kill host cells and phage.
Plating Top Agar with Phage and Host Cells
1. Grow J100265 S1030 host cells overnight in LB broth + 50μg/mL carbenicillin + 10μg/mL tetracycline. After overnight growth, dilute the cells in LB broth + 50μg/mL carbenicillin + 10μg/mL tetracycline to an OD600 of between 0.15 and 0.25. Grow up the diluted cells with shaking and incubation at 37°C until they reach log phase (OD600 between 0.4 and 0.6).
2. While cells are growing to log phase, label bottom agar plates and place them in the incubator in a sealed container for 30 minutes to warm them up. If the plates are moist, they can be left with the lids askew in the incubator for 10 minutes to dry out, and then be transferred to a sealed container.
3. Once cells reach log phase, they are ready for plating. Add 200uL of log phase host cells and 10uL of the diluted phage to a tube of top agar. Vortex the top agar for five seconds, then pour the top agar onto a bottom agar plate. Gently tilt the plate back and forth to evenly spread the top agar. Allow each plate to sit upright on the bench for five minutes before inverting. Plate each top agar tube one by one, so that tubes do not cool too much on the bench before pouring.
4. After allowing the plates five minutes on the bench for the top agar to set, invert the plates and put them into a sealed container in a 37°C incubator. Incubate the plates overnight.
Coring Plaques
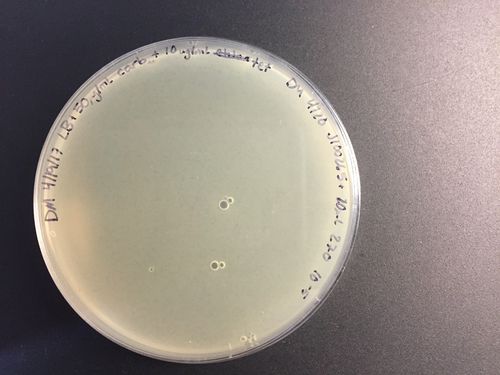
1. After the plates have incubated for 12-18hrs, remove them from the incubator and inspect them for plaques. Plaques are small (~1-3mm diameter) translucent circles. They are best seen when held up to light.
2. Identify a plate which has plaques that are clearly defined and isolated from one another. Prepare multiple 1.5mL microfuge tubes with 1mL of LB broth. Using an inverted 200uL pipette tip, press down on the agar over a plaque to take out a core of agar. Transfer the core into a prepared microfuge tube. Repeat this step for each microfuge tube. Try to keep this process as sterile as possible- for example, use forceps to remove the pipette tip from the box without grabbing the top, which will come in contact with the core.
3. Save the microfuge tubes at 4°C. The phage in the cores will become suspended in the liquid media.
Summary
1. Grow S1030 cells with J100265 and J100270 plasmids overnight to produce SPT7-spec phage. J100269 or J100271 could take the place of the J100270 plasmid for producing promiscuous and wild type phage, respectively.
2. Host cells: Use J100265 cells as host cells. These are the easiest host cells, as geneIII on the J100265 plasmid follows the T7 promoter, and T7 RNAP is produced by the phage genome. J100284 can also be used as host cells, however they must be plated with ATc in top agar or geneIII will not be produced and phage will not be able to produce progeny. J100268 can also be used as host cells, but with the addition of ATc and glucose to repress mutagenesis.
3. Antibiotics: S1030 cells should be grown in or plated with 10ug/mL tetracycline. Bottom agar should have the antibiotic that host cells are resistant to, but not the antibiotic that the phage genome produces resistance to. This is because a phage only confers resistance to a cell that it has infected, at which point the cell’s growth is already inhibited by infection.