Will Notebook2
Wideloache 22:06, 12 March 2009 (EST)
I have forgotten to update my lab notebook since I got back from Spring Break. Here are updates from the past week:
On Sunday night I made new Amp plates for the time delayed growth experiment. I made plates with all combinations of Amp (100 ug/mL, 50 ug/mL. and 25 ug/mL) and Agar (1.5X, 1.0X, 0.5X) - 4 for each combinations - 4 plates for each combination. Plates were given the following number abbreviations:
| 25 ug/mL Amp | 50 ug/mL Amp | 100 ug/mL Amp | |
| 0.5X Agar | 1 | 2 | 3 |
| 1.0X Agar | 4 | 5 | 6 |
| 1.5X Agar | 7 | 8 | 9 |
I was planning on using these plates on Monday-Wednesday for the time delayed growth experiment, however, on Monday I got into Berkeley!!!! and decided to cancel my MIT interview this weekend. That left open the end of this week to do more work in the lab. I decided to do the time delayed growth experiment starting today (Thursday).
Because I had some extra time, I had a chance to get a lower copy Amp plasmid to include in my experiment. The only source of this plasmid (pSB4A5 ~5copies per cell) was from the 2007 registry distribution. This part had BBa_I52001, the cell death gene, as its insert. That meant that I needed to ligate some part into the plasmid before I could transform into cells. I couldn't find any E/P digested inserts that hadn't come from an Amp plasmid in the fridge, therefore on Tuesday I decided to use K091206 (E/P) hoping that the backbone from this digestion was not in the gel purified digestion of the insert. I plated both backbone and insert DNA transformants as controls for this ligation. I also plated this transformation at 25 ug/mL of Amp since pSB4A5 is such a low copy amp plasmid.
On Wednesday I found an approximately equal number of cells on the experimental plate and the insert control plate. There were no cells on the pSB4A5 control plate. This suggested that there was indeed some pSB1A2 backbone in the K091206 GP. Since the ratios were approximately equal (and I needed to start the time delayed growth experiment the next day), I decided to proceed with only high copy number amp colonies. In future experiments, it would be worth testing lower copy amp plasmids as another mechanism to tune the rate of colony growth. I would expect that lower copy amp plasmids, would slow down the rate of colony growth significantly.
Another interesting result from the past week was the sequencing results that I got back on Monday morning. Neither of the sequences for the two promoters contained XbaI or SpeI sites (but they did have EcoRI and PstI sites). This would explain why I have had so much trouble trying to put GFP behind the promoters. The expected length for each promoter part between the EcoRI and PstI sites was 87bp. The pLux' sequence had 104 bp between the E/P sites. The pLuxLas sequence had 88 bp between the E/P site. I had no clue what these two parts could be since they had no homology with the RFP part that I had removed from the pSB1A2 plasmid. I blasted both sequences using NCBI and I got a 100% hit (including the EcoRI and PstI sites) for the pLuxLas sequence in the E coli genome, but no match for the pLux' sequence. Below I have included a screen shot from my blast of pLuxLas:
This result indicated that when I had purified the 87 bp band of pLuxLas from the gel, I probably got some genomic DNA that was also digested with EcoRI and PstI. This genomic insert must have somehow been inserted into the plasmid and been picked through my PCR screen. I have no clue how to explain the pLux' result, other than that some sort of similar, unexpected recombination event must have occured.
After having failed at putting pLux' and pLuxLas into pSB1A2 many many times I have become suspicious that something else must be at play here that is making it difficult to clone these two parts. The iGEM team failed over the summer when they tried primer dimer assembly (they got mutations in all clones). I failed using primer dimer assembly (mutations occured in all cases again) and oligo assembly twice, and I have now failed to move the synthesized promoter from GeneArt into pSB1A2. In the next couple of days I am going to think hard about what could possibly be happening inside the cell that might select against a successful cloning event of pLux' and pLuxLas into pSB1A2. Since these are only promoter parts, it is difficult to imagine a scenario where something like that was happening.
Because it was getting so late in the semester and so close to my thesis being due, I decided to stop trying to clone pLux' and pLuxLas and instead focus on writing. Since I have the transgenic strains compared, I will also test the previously constructed pLas' and pLasLux promoters for fluorescence in an experiment tomorrow.
Wideloache 16:56, 28 February 2009 (EST)
I was really busy yesterday, so I wasn't able to come in a do the transformations. Today I miniprepped S03981 and S03984. I then performed a TSS transformation of these two preps with MC4100::K091206. I plated on amp plates (although I could have, and maybe should have, plated on amp+gen). I put these plates on the benchtop for growth over two nights. I will ask Dr. Campbell to put them in the fridge for when I get back from spring break. I will streak these cells on Gen plates to make sure that they still have the genomic insert (once I do this I will be completely convinced of the genomic insertion having worked) and then test for fluoresence when provided with the proper autoinducer molecules (in the -4C freezer).
I also decided to have pSB1A2-pLux and pSB1A2-pLuxLas sequenced over the break. I figured this would help me determine why SpeI wasn't cutting. I prepared the samples and left them on Dr. Campbell's desk so that he could ship them sometime next week. Once I get back I will need to move forward with testing the pLas promoters, ligate GFP behind the pLux promoters, and test the time delayed growth for varying amp/agar concentrations. Hopefully I can get all of this done before my thesis is due, but traveling frequently this semester has slowed me down.
Wideloache 14:22, 26 February 2009 (EST)
I realized today that I didn't have minipreps for S03984 and S03981 so I picked them out of the -80 (iGEM 2008 box). I will do a TSS transformation of these minipreps tomorrow.
Wideloache 18:01, 25 February 2009 (EST)
I am back from Berkeley (came in on a redeye this morning). In lab meeting today I had a though about the unexpected PCR results for pG80ko-K091206 with the 4 integration primers. Assuming that the plasmid prep wasn't totally clean, I could have JM109 genomic DNA in the sample. If this was the case, then I would expect a band for primer 1/4. This explains the band in lane 9, but lanes 10 and 11 are still puzzling.
I grew up some MC4100::K091206 in preparation for transforming in pLasLux-GFP (BBa_S03984) and pLas'-GFP (BBa_S03981). Since I can't seem to get the ligation of pLux and pLuxLas to work, I will just proceed with the prebuilt promoters to see if the integrant cell strains works.
Wideloache 17:23, 20 February 2009 (EST)
I took my transformation plates out of the incubator and found that, while I didn't have a lawn, I still had lots of colonies on all of the plates, including negative control. Based on this result, I think that the pLux and pLuxLas are perhaps not cutting with SpeI. I had suspected this before, but then decided that maybe the bad amp was the cause of the high background. If SpeI wasn't cutting, then the colonies that I have on the plate (post SAP procedure) were the ones that didn't cut with PstI either. This seems to be the most likely explanation. I will investigate this possibility when I get back from Berkeley, but I will be gone until Wednesday at an interview.
Wideloache 19:04, 19 February 2009 (EST)
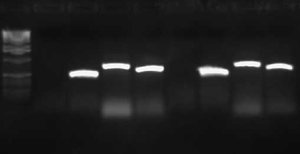
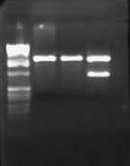
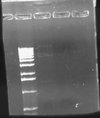
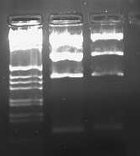
I wasn't able to run Tuesday's PCR on a gel yesterday because I was working on an artificial intelligence independent study project all day. Today, though, I ran the PCR (MC4100:K091206 with controls) on a 1.0 gel and obtained the following results:
These gel results seem to confirm the genomic integration. The results for MC4100::K091206 were the same results as Tuesday, where a band was not seen for 1/4 but it was for all other primer combinations. This suggests multiple integrants exist in the strain. Additionally, this gel confirms that MC4100 cells do give a band for primers 1/4 (but not for any of the other primer combinations). This is explained because primers 1 and 4 face each other in the wildtype but are pushed 4000bps farther apart in the integrant strain. The unexpected result from this gel was the pG80ko-K091206 results. While I did get the band from 3/2 as expected, I did not expect to see a band for any of the other lanes. This could be because of regions of homology existing between the primers and the plasmid. I will investigate this possibility further in the next couple of days, but since the 3/2 band is brighter than the others I might be able to raise the annealing temperature and get rid of those bands.
Today I also redid the ligation/transformation of pLux and pLuxLas with E0240 (GFP). I used the same SAP treated pLux and pLuxLas samples that I had used earlier this week on the bad plates, but I plated on new Amp plates that Kin had made. Hopefully the SAP procedure will get rid of the background.
Wideloache 17:32, 18 February 2009 (EST)
My transformation of pLux' and pLuxLas + GFP gave a lawn on all 4 plates. This suggested that the amp in the fridge was bad (since Kin made new plates yesterday after finding that the amp plates in the fridge gave a lawn). We threw away the bad amp. This also explains why all of the colonies came up at the same time when I was trying to do the time delayed growth experiment. The amp on all of those plates in bad as well, which means I will have to remake all 9 combinations of agar and amp. I will have to redo the transformation of pLux and pLuxLas + GFP once we make new amp plates.
Wideloache 16:56, 17 February 2009 (EST)
I ran a PCR screen for successful genomic insertion using the two cultures that I had grown up overnight. I used the primers that I had ordered from earlier. As a reminder, those sequences were:
attPhi80-1: CTGCTTGTGGTGGTGAAT
attPhi80-2: ACTTAACGGCTGACATGG
attPhi80-3: ACGAGTATCGAGATGGCA
attPhi80-4: TAAGGCAAGACGATCAGG
The expected results are shown in the table below. Primer sequences and expected results are based on the CRIM paper found here. As specified in the paper I used an 10 min denaturation @ 94C followed by 25 cycles of 1 min @ 94, 1 min @ 63, 1 min @ 72.
| sequence Temp (°C) | No integrant with 1 and 4 | Single integrant with 1 and 2, 3 and 4 | Multiple integrant with 1 and 2, 3 and 2, 3 and 4 |
| 63 | 546 | 409, 732 | 409, 595, 732 |
Based on these results it appears that I finally got the genomic insertion to work correctly!!! It looks like I have multiple integrants in each of the clones as the band lengths matched with the expected bands for PCR with multiple integrants. This may be a good thing given the fact that have the expression cassette on the genome will significantly reduce the expression of LuxR and LasR.
From now on I will use clone 2 as my integrated cell strain. It will be referred to as MC4100::K091206. I needed to redo this PCR procedure with MC4100::K091206 using normal MC4100 cells and pG80ko-K091206 as controls. I ran this PCR O/N using the same primer combinations with MC4100:K091206 and the two controls (pG80ko-K091206 miniprep and MC4100+pInt80-649 cells). I will run this PCR on a gel tomorrow.
I also did a shrimp alkaline phosphatase procedure on the pLux' and pLuxLas digestions (S/P) before ligating with E0240 (X/P). I used buffer low, which Samantha said worked for her (all of the SAP buffer was gone). Hopefully this procedure will remove all background, and I can see if any of the promoter plasmids were able to digest with SpeI. If I get no colonies on these plates, then I may have to send the promoters off for sequencing at the SpeI site.
Finally, I noticed that the computer had run out of hard drive space after taking 54 time lapse images. I stopped the imaging because I also noticed that the Ec100D:pir+ cells had come up at the same time as the amp colonies. This suggested that maybe the amp concentration of that plates wasn't high enough to prevent growth of Ec100D::pir+. I streaked more cells on the plate and grew it up at 37C O/N. I am now thinking that testing all plates in parallel might be a better option. I could come in every few hours and take images of all of the plates. This might give me enough data for the model, and I would be able to use all 18 combinations. I will present this idea at lab meeting tomorrow. Maybe I could sleep in the lab for one night when I did this?
Wideloache 14:08, 16 February 2009 (EST)
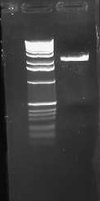
Today I ran my digestion of pLux' and pLuxLas with EcoRI and SpeI on a 2.0% gel and obtained the following result:
Neither band appeared to be present in the gel image. This suggests that possibly lack of cutting at the SpeI has been the cause of my high negative control background on my transformations. I will need to do PR and possibly sequencing of the parts to investigate further what is going on.
I also began the first time delayed growth measurement. I used pSB1A2 (pSB1A2-pLux') as my amp producing colony. I used Ec100D::pir+ streaked from liquid culture as my non amp resistant cells. And I used an Amp plate with 0.5X agar and 50ug/mL of amp.
Wideloache 21:53, 14 February 2009 (EST)
I came in and found that there were colonies on my LB+Gen plate of MC4100+pInt80-649+pG80ko-K091206 at 43C!!!!!
I put this plate in the fridge and will run a colony PCR on it after the weekend (busy with an ultimate tournament this weekend).
Wideloache 23:25, 13 February 2009 (EST)
I ran my digestion of pLux', pLuxLas, and the digestion control (J23100-J61002) on a 1.0% gel:
The control band was as expected for a successful digestion, suggesting that the SpeI enzyme is not the cause of the high background. I excised these bands and saved them in the fridge for gel purification and alkaline phosphotase tomorrow. I realized that it is possible that maybe the DNA sent from GeneArt doesn't have a proper SpeI site, or methylation of the SpeI site is occuring, or the parts aren't actually in the plasmid as I expected them to be. To make sure that SpeI digestion can occur in these plasmids, I ran an O/N digestion of both promoter parts with EcoRI and SpeI. If the promoters are cut out by this digestion, then I will perform alkaline phosphotase ligations of the two parts with E0240. If not, then I will have to investigate what is preventing cutting with SpeI.
I also noted that I had 6 colonies on the transformation plate of MC4100+pInt80-649+pG80ko-K091206. I had no colonies on the corresponding HB101 plate:
This might suggest that HB101 has some problem with maintaining one or both of these plasmids at the same time. This was positive evidence that maybe the cell strain was the cause of things not working. I grew a colony from the MC4100 plate up in LB+Gen and in LB+Gen+Amp. I let it grow for about 6 hours and then streaked from teh LB+Gen culture onto a LB+Gen plate. This culture appeared to be growing faster (perhaps because more cells were added to the culure initially, but perhaps because many of the cells had the genomic insertion already and the integration plasmid wasn't happy at 37C??). Anyway I put the LB+Gen plate at 43C for O/N growth. Tomorrow, if there are no colonies, I will try again with the procedure from the paper, where I shock the cells at 43C for 30 minutes and then plate at 37C.
I also made Amp plates in preparation for modeling the beta lactamase diffusion which leads to time delayed cell growth. I made 9 types of plates with all combinations of 3 different amp concentrations and 3 different agar concentrations. The amp concentrations were: 100 ug/mL, 50 ug/mL, and 25 ug/mL. The agar concentrations were: 0.5X normal agar (7.5g/liter), 1.0X normal agar (15g/liter), and 1.5X normal agar (22.5g/liter). I will also be using two different copy number plasmids for the amp production (pSB1A2 and pSB4A5). I will begin testing these tomorrow by growing pSB1A2 producing cells as the amp producer and Ec100D:pir+ cells as the time delayed growers (same as last time). I will perform each growth in triplicate (three streaks per plate measured). The streaks will be continuous so that I can get a continuous distribution of growth distances. I grew up Ec100D::pir+ cells in liquid LB O/N so that I could use them for streaking tomorrow. I will always streak from liquid so that I am consistent with the number of cells on the plate.
Wideloache 18:20, 11 February 2009 (EST)
Today I yet again had extremely high background on my negative control transformation plate. I still have no GFP on the first set of transformation plates. I will continue to monitor both and leave them on the bench, but I am doubtful that either will contain fluorescent colonies. It is possible that SpeI is not cutting the plasmid. I will do a third digestion of pLux and pLuxLas with Spe/Pst and use J23100-J61002 as a control for cutting with Spe and Pst (This plasmid contains an RFP insert between the SpeI and PstI sites). I will run this on a gel and, assuming that SpeI has cut the control, I will do a Alkaline Phosphotase procedure on the plasmid to reduce background. I began this digestion and ran it O/N.
I also did a TSS transformation of pG80ko-K091206 into HB101+pInt80-649 and MC4100+pInt80-649. I rescued for about 30 minutes at room temp and about 30 minutes at 30C before plating on LB+Gen plates. If I get colonies tomorrow I will grow them up at 37 and streak at 43C.
Wideloache 16:08, 10 February 2009 (EST)
Today I pulled my MC4100 + pInt80-649 transformation plate out of the 30C incubator and found that I had two colonies. I expected a higher yield, but I picked one of these colonies for O/N growth at 30C in LB+Amp. I also grew up HB101+pInt80-649 in LB+Amp at 30C. Tomorrow I will do a TSS transformation of both with pG80ko-K091206.
I also ran my digestion of pLux', pLuxLas, and E0240 (GFP) on a 1.0% gel. This time I was careful to use TAE buffer for my gel instead of water!
My gel ran as expected. I gel purified the two vectors from lanes 1 and 2 and the insert from lane 3. I did ligations of pLux' and pLuxLas to E0240 as I had done two days ago, transformed into zyppy cells, and plated on LB+Amp plates.
Wideloache 23:56, 9 February 2009 (EST)
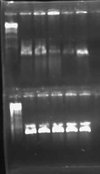
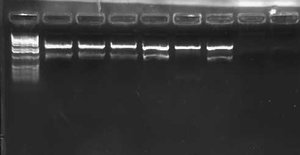
I had an excessive amount of background colonies on my transformation plates for both pLux' and pLuxLas. I did a colony PCR of 5 colonies from each experimental plate and obtained the following result:
My gel was extremely smeared because I made it with water instead of TAE buffer. WOW! Rookie mistake. Anyway, I could still tell based on this screen that no colonies had the GFP insert. A successful product would have been about 1000 bps while an unsuccessful ligation would have given bands of approximately 250 bps. I will leave the plates on the bench O/N to see if any colonies fluoresce green. This is unlikely, however, since the promoter is supposed to be constitutively off.
I also re-digested pLux', pLuxLas, and E0240 O/N to see if I could get a better yield by going through the process again.
Wideloache 18:16, 8 February 2009 (EST)
Today I ran the digestions on a 1.0% gel.
These results show that pLux'-1 probably does not contain a successful ligation. Perhaps it gave a band length because of a funny ligation event. In any case, pLux'-7 was confirmed to have a successful insert in it (~100bps). I purfied the (S/P) digestion from Lane 4 and used this DNA for a ligation with E0240 (X/P). I verified the length of the E0240 GP by running it on the gel. I had found this tube in the freezer and verified its contents from James' lab notebook.
I had forgotten to digest pLuxLas with S/P on Friday, so I began another digestion and let it run for ~2 hours. I ran this on a 1.0% gel:
I gel purified this band and ligated to E0240. I zyppy transformed both the pSB1A2-pLux'-GFP-TT and pSB1A2-pLuxLas-GFP-TT parts into JM109 cells.
I also grew MC4100 up and performed a TSS transformation of pInt80-649 into the cells. The pInt80-649 digestion from the first gel above gave a band at approximately the expected length (1400) but not at the expected length for the larger band (5000). There may have been way too much DNA in the miniprep since I used 16uL of DNA. Perhaps growing the cells at 30C instead of 37C had a significant effect on the miniprep efficiency. I plated the transformed cells on an LB+Amp plate at 30C after a 1 hour rescue at 37C (oops, forgot to rescue at 30C).
Wideloache 14:22, 6 February 2009 (EST)
Yesterday, I grew up HB101+pInt80-649 cells in LB+Amp O/N at 30C. Today I miniprepped these cells. I am hoping that the lower temperature kept plasmid happier, and I will get a better yield from my miniprep. I digested the pInt80-649 miniprep with EcoRI and XbaI. This digestion should yield band lengths of 1500 and 5000 bps. I also miniprepped my samples of pLux' (clones 1 and 7). I did an O/N digestion of each with S/P and E/P. The S/P digestion will be used for ligation with E0240 (X/P). The E/P digestion will be used to verify the band length.
Wideloache 17:29, 4 February 2009 (EST)
I ran my O/N digestion of pInt80-649 on a gel and got the following result:
Even though I had used 15uL of DNA, the bands were still too faint to see. I will try growing the HB101+pInt80-649 cells at 30C O/N before miniprepping them. Perhaps they weren't happy at 37C and didn't give enough product. If this is really how low the copy number is of this plasmid, then it is going to be difficult to verify through a gel. Tomorrow I will investigate the possibility of PCR using the primers I ordered to verify the plasmid.
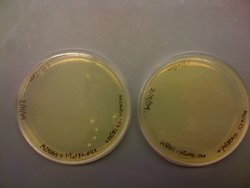
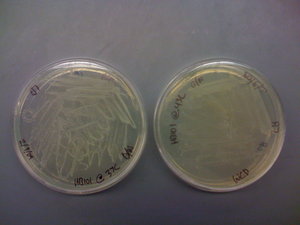
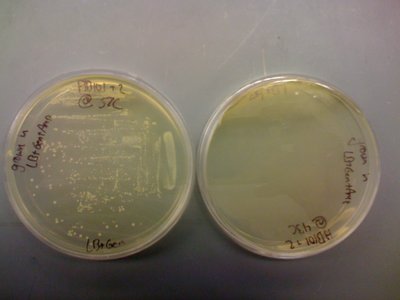
I also found that the HB101 cells did grow at 43C. This means that temperature is likely not the problem. I included pictures of the plates below:
It should be noted that there was less growth on the 43C plate, but the fact that there was growth means that the cells should still be able to grow O/N. Regardless, I will try the heat shock at 43C protocol that was described in the CRIM paper before eliminating temperature as a possible source of this not working.
Wideloache 23:57, 3 February 2009 (EST)
In my meeting with Dr. Campbell, we went over the possible things that could be going wrong with the genomic insertion into HB101. The two main possibilities that we came up with were that HB101 couldn't grow at 43C or that the plasmid sent from Chris was wrong. To test whether HB101's can grow at 43C, I plated some cells on an LB plate at 43C and at 37C for O/N growth. I found in the CRIM paper that they suggested at heat shock at 43C for 30 minutes instead of full growth O/N. Chris' email simply said to "plate at 43." It didn't say for how long, so its possible that the exposure at 43C is only supposed to be for a short time. Writing this, I'm not getting more confident that this is the problem. Last semester I kept a plate that had been at 43C O/N out on the bench and got colonies to come up. To test whether these colonies had the genomic insertion, I tried to grow them at 43C and they didn't. It is totally possible that they were successful genomic insertions that simply couldn't grow at 43C. If this is the case, then finishing up the genomic insertion will be fairly straight forward. I will have to wait until tomorrow for these results. In the meantime, I also digested a higher concentration of pInt80-649 (16uL of DNA) with EcoRI and XbaI. If the HB101's do grow at 43C, I will run this digestion on a gel to see if the plasmid sent by Chris gives the expected bands at 1500bps and 4000bps.
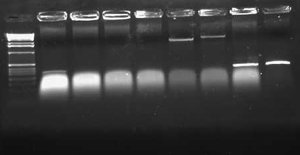
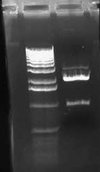
I also did a colony PCR of the 7 remaining pLux' colonies from my transformation last week. I included a positive control of the pLuxLas cells that had been confirmed yesterday (clone #2). I obtained the following result:
This gel suggests that clone #1 and #7 probably have the pLux' insert. The reason that lane 8 has no junk at the bottom is probably that more cells were in the reaction since I took the positive control from liquid culture and not a plate. I grew both clone 1 and 7 up O/N and will miniprep them tomorrow. At that point, I will do an analytic digest on them to further confirm successful ligation. If they are correct, I will proceed with ligation of pLux' and pLuxLas to GFP-TT (E0240), which I found a GP of in the freezer.
Wideloache 23:44, 2 February 2009 (EST)
Dr. Campbell found MC4100 in the freezer. I picked cells from the tube and grew them up O/N in LB. I also moved the tube into my freezer stocks box and added it to the database (Box 4-1 #35).
I found that I had growth for 2 out of the 3 cultures containing HB101+pInt80-649. The culture from the freezer didn't grow (again), but the cells from the plate in the fridge and the tube from Wed did grow. I miniprepped these cells and digested with EcoRI and XbaI. This should give bands at 4,000 and 1,500. It should also be noted that the concentration of my minipreps were ~10 ng/uL. This suggests that maybe the plasmids are not totally happy at 37C. (I'm not sure what the copy number for the plasmid is, but I would imagine it should give a higher miniprep concentration even if it was a low copy plasmid.
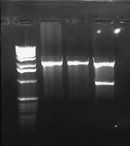
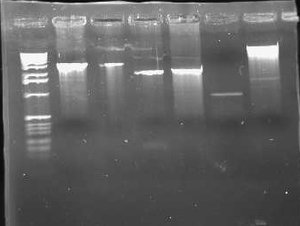
I also miniprepped the 3 cultures of pLux' and pLuxLas that were picked from the transformation plates yesterday. I digested these with EcoRI and PstI and ran it on a 0.8% gel. A successful ligation should give bands that are a little >100bps. I also ran my digestions of pInt80-649 on the gel.
Based on this gel, it appeared that only one of the 6 Lux promoter constructs was correct (pLuxLas clone 2 in Lane 5.) The others gave inserts that were way too large (>1000). I didn't run the gel any farther because I already had the information I needed. I also found that I need to use much more DNA (I used 5 uL) for the pInt80-649 digestions (because of the low miniprep concentrations). The bands were barely visible to the point that they weren't very helpful. Since I had to go, I decided to redigest pInt80-649 tomorrow at a higher concentration and run it on another gel. Based on a guess, though, it appeared that a band did likely exist at about 1500bps or thereabouts. I couldn't make out any second band above it though.
Run colony PCR on all pLux' colonies. Pick pLas'-GFP-TT and pLasLux-GFP-TT out of the freezer Ligate GFP-TT (E0240) behind pLuxLas. (I found a E0240 MP in the freezer) Digest pInt80-649 with E/X and run on gel. If verified pInt80-649, TSS transform into MC4100
Wideloache 23:21, 1 February 2009 (EST)
I returned from Harvard this afternoon and found that Dr. Campbell had put my transformation plates in the fridge. There were satellite colonies around most of the colonies, but many of the colonies on the negative control plate (but not all) had become red. There were some red colonies on the experimental plates, although the ratio of white:red was lower than it was on the negative control. This suggested that the ligations may have worked. I picked 3 colonies of both pLux' and pLuxLas for O/N growth in LB+Amp. Tomorrow I will miniprep these cultures to verify successful ligation.
I also found that the culture of HB101+pInt80-649 (which, after not growing Wed night, I had left out on the bench over the weekend) had a good bit of growth in it. I picked new HB101+pInt80-649 cells from the freezer, cells from the tube with growth, and cells from an old plate in the fridge for O/N growth in LB+Amp. I will see what of these grows in the culture. Whatever does grow should contain the integration plasmid, which I will miniprep and transform into MC4100 (which I still need to find in the freezer).
Wideloache 12:05, 29 January 2009 (EST)
I am leaving to interview at Harvard this afternoon, but I came to check on my transformations from yesterday. They were pretty slow growing, and none of the colonies appeared red. There were 5-10 colonies on the negative control and a similar number on the other 4 plates. I asked Dr. Campbell to take them out of the incubator after the red color started to appear (as I expect it will in some colonies).
I also surprisingly had no growth in my tube of HB101+pInt80-649. I will pick more cells from the freezer and also from the fridge (where I have an old plate), once I get back from Harvard.
Wideloache 19:37, 28 January 2009 (EST)
I had no growth on any of the plates from my Monday afternoon transformations. Initially, I suspected that the ligase I was using was bad. However, during lab meeting today I became aware of the fact that Samantha had been doing heat shock transformations with the JM109 cells. It turns out that we have two varieties of JM109 cells in the freezer, only some of them are zyppy cells and the others require heat shock. This explains why none of my stuff has been working for a long time. The transformation efficiency has been really low because the cells I have been using aren't zyppy cells, and I haven't been doing heat shock transformations. Therefore, I redid the ligations of pSB1A2 (newer digestion) to pLux and pLuxLas (both the older and newer digestions). I transformed all 4 of these ligations and a control ligation (pSB1A2 only) into JM109 ZYPPY CELLS!!!!! and plated on LB+Amp plates.
Additionally, in lab meeting Dr. Campbell posed the idea that HB101 cells didn't have the attPhi80 sites in their genome, which would explain why the genomic insertion wasn't working in that cell strain. I looked through the literature on HB101 cells and found the following description:
Genotype: F-∆(gpt-proA)62 leuB6 supE44 ara-14 galkK2 lacY1 ∆(mcrC-mrr) rpsL20 (Strr ) xyl-5 mtl-1 recA13(4)
I don't know what any of that means, but none of it appears to deal with attPhi80 sites. I will ask Dr. Campbell to take a look at some point, but for now I will continue with the genomic insertion into MC4100. I looked through the freezer to find Andrew's old stock of MC4100, but I couldn't find it anywhere. I will also ask Dr. Campbell about this tomorrow.
Finally, I picked HB101 cells containing pInt80-649 from the freezer and grew them up at 37C O/N in LB+Amp.
Wideloache 15:14, 26 January 2009 (EST)
Once again, I had growth on the LB+Gen(10ug/mL) plate at 37C, but not at 43C. I took a picture to document these results:
The only possible reason for this failing that I can think of is that the HB101 cells are not capable of undergoing this genomic insertion. I will try 2 things:
- Streak normal HB101 cells and grow them at 43C to ensure they are capable of growth at that temperature
- Try the genomic insertion in MC4100 cells in parallel with HB101 cells.
I also ran the E/P digestion of pLux' and pLuxLas on a 2.0% gel:
The expected length of the bands was 93bps. I excised the lowest bands in each of the lanes, did a gel purification, ligated each with pSB1A2, and transformed into JM109 cells.
I also transformed pInt80-649 into JM109 cells. I was out of DNA, but I still had the tube Chris had sent, so I added 20 uL of H20 to the tube and then transformed in 50 uL of Zyppy cells. I also have some HB101 +pInt80-649 cells in the -80 freezer. I didn't use these to get the pInt80-649 because I wanted to start as far back as I could go. Using the HB101 cells to get more pInt80-649 would have left me wondering if that was the correct DNA if things didn't work. If I get cells from the transformation, I will pick one and grow it up O/N.
Wideloache 17:45, 25 January 2009 (EST)
Both colonies from the transformation (which I had grown up in LB+Amp O/N) were fluorescing red in the culture, meaning that the ligation had been unsuccessful. I will have to try this assembly a 3rd time!!!! I redigested pLux and pLuxLas O/N from the stock sent from GeneArt. I suspect that maybe PstI didn't cut in the digestions of either pSB1A2 or pLux' and pLuxLas. This would explain why pSB1A2 only gave one band (although that band appeared to be the correct size for the vector.) The difference in size for pLux' and pLuxLas when digested with EcoRI and EcoRI/PstI is 7bps and pretty much indistinguishable.
I also had growth on the 2 LB+Gen plates at 37C (one of which had HB101+pInt80-649+pG80ko-K091206 cells grown in LB+Gen liquid culture and the other in LB+Gen+Amp). There were significantly more colonies on the plate which had colonies from LB+Gen+Amp than there were from the liquid culture of LB+Gen. There were no colonies on either of the plates which were grown at 43C. I am going to try this experiment again, this time using LB+Gen plates at 10ug/mL instead of 20 ug/mL (and only colonies that grew in LB+Gen+Amp). This will tell me if the concentration of gen is too high on the plates for a successful genomic integration event to grow.
Wideloache 13:19, 24 January 2009 (EST)
I noted that the HB101+pInt80-649+pG80ko-K091206 cells from Thursday had grown in LB+Gen and in LB+Gen+Amp after 1 day. There was, however, no growth in LB+Amp alone after 2 days. I suspect that the colony I picked for this culture was too small and that I hadn't gotten any cells into the media. I moved some cells from the LB+Gen+Amp liquid culture into LB+Amp culture to ensure that growth could occur.
Since the cells in the LB+Gen and LB+Gen+Amp culture appeared to have both integration plasmids, I streaked them on plates to attempt the genomic integration. I first streaked cells from the LB+Gen culture and the LB+Gen+Amp culture onto LB+Gen plates (20 ug/mL) at 37C. I did the same thing at 43C. If the cells grow at 37C and not at 43C, then something is probably going wrong with the integration event. If they don't grow at 37C then I will be really confused.
For my transformation of pSB1A2-pLux' and pSB1A2-pLuxLas, I had one colony on each of my experimental plates, but no colonies on my control plate. Both colonies appeared red, but I picked them anyway and grew them up O/N in LB+Amp.
Wideloache 15:40, 23 January 2009 (EST)
Today I did the gel purification of pSB1A2 (E/P). I then ligated this vector with the previously digested pLux' and pLuxLas promoters and transformed into JM109 cells. The unsuccessful ligation products should yield red colonies, since RFP was cut out of pSB1A2. Successful ligations should give non-fluorescent colonies.
Wideloache 14:25, 22 January 2009 (EST)
After two days, there was still no growth in the 2 liquid cultures (Amp/Gen and Gen). There were 5-6 colonies on each of the LB + Gen plates (one was at 10 ug/mL and one was at 20 ug/mL). I streaked one of the colonies onto an LB + Gen plate at 20 ug/mL to save in the fridge. I also grew up 3 colonies in different liquid culture conditions at 37C: LB+Gen, LB+Gen+Amp, and LB+Amp. This should give me information on whether both plasmids are in the cells. Cells that have the genomics insertion only should only grow in LB+Gen. Cells that have both of the insertion plasmids should grow in all 3 conditions.
I also miniprepped and digested J61002-J23100 with EcoRI and PstI (45 min). This should give me pSB1A2 vector with RFP cut out. I ran this digestion on a 0.8% gel and obtained the following result:
I excised the bright band in the middle of the 3 bands in lane 1. This appeared to be the doubly digested vector (~2000 bps). Singly cut plasmid likely accounted for the top band and the insert appears at the bottom of the lane (~700 bps).
Wideloache 17:14, 20 January 2009 (EST)
On the 16th, I found that my negative control transformation had tons of colonies (almost a lawn). The two experimental plates had equal numbers of colonies. I didn't bother testing these colonies because they were almost certainly not the desired constructs. I'm going to have to redo the transformation with a new vector digestion. Therefore, today I grew up J61002-J23100 in LB + Amp. Tomorrow I will miniprep and digest the plasmid in preparation for another transformation. It is possible that the MP of J61002-J23100 was bad, since I didn't have the second expected band when I did the E/P digestion on the 15th.
I also made new Gentamicin plates and LB + Gen liquid culture. I made two different plate concentrations: 10 ug/mL and 20 ug/mL. My liquid culture was at 15 ug/mL which was the concentration in which Chris said to grow up the colonies containing pInt80 and pG80ko. [Note: Last semester I had used 20 ug/mL, which was the literature working concentration. When I did the procedure last semester I figured that the concentration difference wouldn't be a problem, but since I'm trouble shooting now, I made new liquid culture at the correct concentration.]
I picked HB101 +pInt80-649 +pG80ko-K091206 out of the -80. I streaked them out of the LB Gen plates at both concentrations. I will keep track of the growth of these cells to see if there is a difference in growth time. If the 10 ug/mL plates have faster growing cells, this might suggest that the gentamicin concentration of 20 ug/mL was too high. I grew these plates at 37.
I grew the HB101 +pInt80-649 +pG80ko-K091206 cells in LB + gen and LB +gen+amp liquid culture at 37. Theoretically these cells should grow in both tubes.
At this point, most of these variable growth conditions are just to try to wrap my head around what could be going on inside the cells based on their response to different growth conditions.
Wideloache 15:45, 15 January 2009 (EST)
I ran my digestion of J61002-J23100 (E/P) on a 0.8% gel and got the following result:
I expected to see a band at around 900, which would have been the RFP insert, but since the vector was fairly faint as well, it probably just wasn't bright enough to see. I excised the band at around 2000 bps which should be pSB1A2 vector cut with E/P.
I also ran my digestions of LacIX86-pLux' and LacII12X86-pLuxLas on a 2.0% gel and got the following result.
This was what I expected to see, and I excised the lowest band on each gel (expected size was 93 bps).
I did a gel purification of all 3 excised gel fragments, and then I ligated pSB1A2 with each of the pLux promoter parts. I also did a negative control ligation of pSB1A2 + H20. I expect to have the unsuccessful ligation be red, making the verification of successful ligations easier. I plated on LB Amp plates after transforming into JM109 cells.
Wideloache 20:36, 13 January 2009 (EST)
Today was my first day back in the lab of the semester. The first thing that I needed to do was to get the two pLux promoters that had been synthesized by GeneArt into pSB1A2. I had done an (E/P) digestion of pLux' and pLuxLas last semester that seemed to be successful. However, when I tried to transform into pSB1A2, I had a low colony yield. I decided to make a fresh stock of pSB1A2 digested with E/P. I used the part J61002-J23100. Cutting this with E/P woudl result in a pSB1A2 vector with a RFP expression insert. In this way I could screen out unsuccessful ligations. In addition to running this digestion O/N, I also digested pLux' and pLuxLas with E/P from the minipreps of LacIX86-pLux' and LacII12X86-pLuxLas.
Make GenR plates (15 ug/mL) Miniprep pLas' and pLasLux Get GFP plasmid to put 4 promoters in front of.