Difference between revisions of "Eukaryotic Models"
| Line 1: | Line 1: | ||
=Eukaryotic Models of Stochasticity in Gene Expression= | =Eukaryotic Models of Stochasticity in Gene Expression= | ||
Eukaryotic Models of stochasticity differ from prokaryotic models becuase their processes of transcription and translation are inherently different. Therefore, in stochastic models, different single variables are needed to account for discrepencies. Collins et. al. created a model for Stochasticity in Eukaryotes, focusing on the stabilization of the transcriptional complex to mRNA. Where states PC1, PC2, and PC3 all detail different states of DNA. PC1 represents the inactive DNA, PC2 represents an intermediate complex where TATA binding protein and other transcriptional factors are bound, and PC3 represents the preinitiation complex. Another characterization of Eukaryotic expression lies in the alpha variable which represents masking and unmasking of DNA by chromatin structure (Collins et. al., 2003). | Eukaryotic Models of stochasticity differ from prokaryotic models becuase their processes of transcription and translation are inherently different. Therefore, in stochastic models, different single variables are needed to account for discrepencies. Collins et. al. created a model for Stochasticity in Eukaryotes, focusing on the stabilization of the transcriptional complex to mRNA. Where states PC1, PC2, and PC3 all detail different states of DNA. PC1 represents the inactive DNA, PC2 represents an intermediate complex where TATA binding protein and other transcriptional factors are bound, and PC3 represents the preinitiation complex. Another characterization of Eukaryotic expression lies in the alpha variable which represents masking and unmasking of DNA by chromatin structure (Collins et. al., 2003). | ||
| − | + | <br><br> | |
| − | + | Figure 1 | |
| − | [[Image:Eukaryotic_stochastic_model.jpg | + | [[Image:400px-Eukaryotic_stochastic_model.jpg ]] |
| + | <br> | ||
| + | Figure 1 was obtained at | ||
<center> [[Modeling Stochasticity | Back to Modeling Stochasticity]]</center> | <center> [[Modeling Stochasticity | Back to Modeling Stochasticity]]</center> | ||
Revision as of 02:38, 6 December 2007
Eukaryotic Models of Stochasticity in Gene Expression
Eukaryotic Models of stochasticity differ from prokaryotic models becuase their processes of transcription and translation are inherently different. Therefore, in stochastic models, different single variables are needed to account for discrepencies. Collins et. al. created a model for Stochasticity in Eukaryotes, focusing on the stabilization of the transcriptional complex to mRNA. Where states PC1, PC2, and PC3 all detail different states of DNA. PC1 represents the inactive DNA, PC2 represents an intermediate complex where TATA binding protein and other transcriptional factors are bound, and PC3 represents the preinitiation complex. Another characterization of Eukaryotic expression lies in the alpha variable which represents masking and unmasking of DNA by chromatin structure (Collins et. al., 2003).
Figure 1
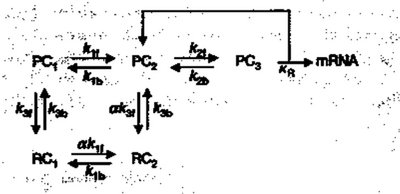
Figure 1 was obtained at